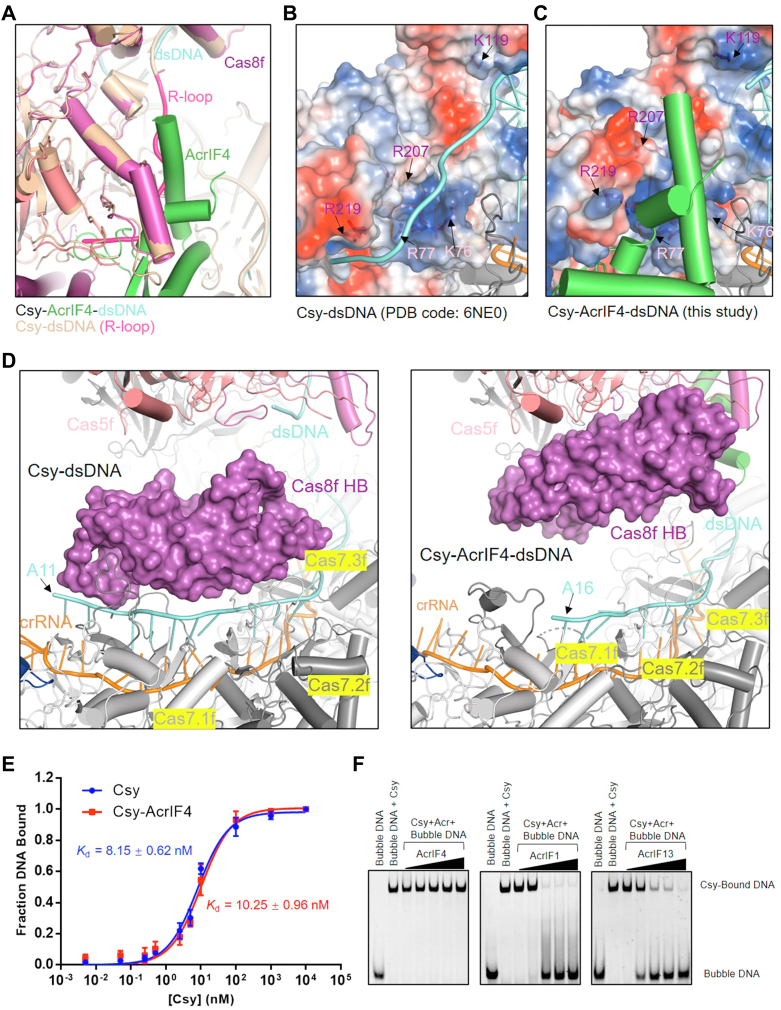
Figure 3.
AcrIF4 binding to Csy destabilizes the R-loop formed by the NTS.A, a close-up view of the R-loop region of the superimposition of the structures of the Csy-AcrIF4-dsDNA and Csy–dsDNA. The R-loop in Csy-dsDNA is colored in hot pink. B and C, R-loop binding channel (RBC) in the structure of Csy-dsDNA (B) and Csy-AcrIF4-dsDNA (C). The Cas8f and Cas5f residues lining the RBC are shown as sticks and colored as the subunits in Figure 2A. The Cas8f and Cas5f subunits in two structures are shown in electrostatic model. D, a close-up view of the 5′ end of TS in the Csy-dsDNA (top) and Csy-AcrIF4-dsDNA (bottom) complexes. The Cas8f HB in two structures are shown in surface model. E, the binding affinities of “bubble” dsDNA by the apo Csy or the Csy–AcrIF4 complex tested by EMSA. Error bars represent SEM; n = 3. The KD values are 8.15 ± 0.62 and 10.25 ± 0.96 nM for the Csy and Csy-AcrIF4, respectively. Raw data for these curves are shown in Fig. S5. F, EMSA assays showing that incubation with AcrIF4 has no effect on the binding of “bubble” DNA. Reactions were performed with 1.6 μM Csy, 0.1 μM “bubble” dsDNA and Acr concentrations of 0.4, 1.6, 6.4, 25.6, and 102.4 μM following the order indicated by the black triangle. Csy, crRNA-guided surveillance; EMSA, electrophoretic mobility shift assay; NTS, nontarget strand.