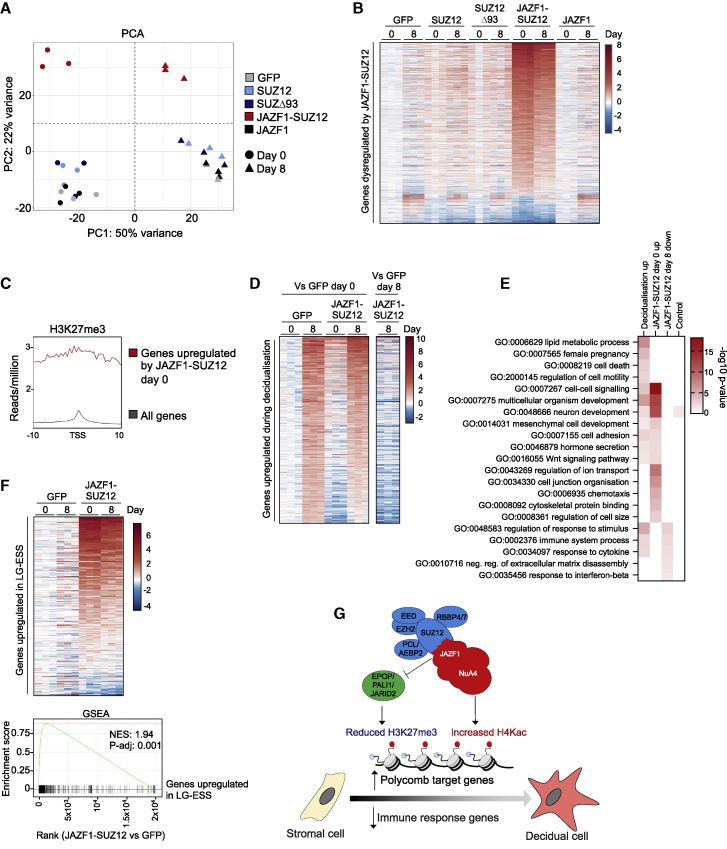
Figure 5.
JAZF1-SUZ12 activates polycomb target genes and dysregulates decidualization in primary hEnSC
(A) Principal component analysis of RNA-seq data from undifferentiated (day 0) and decidualized (day 8) hEnSCs from three donors expressing GFP, SUZ12, SUZ12Δ93, JAZF1-SUZ12, or JAZF1.
(B) Genes (n = 909) that exhibit significant changes in expression in hEnSCs containing JAZF1-SUZ12 at day 0 or 8. The colors indicate log2 fold change in expression of these genes in hEnSCs from three donors containing GFP, SUZ12, SUZ12Δ93, JAZF1-SUZ12, or JAZF1 relative to the average expression in hEnSCs containing GFP at day 0, according to the scale on the right.
(C) Metagene plot showing average H3K27me3 occupancy in hEnSCs around the transcription start site (TSS) of genes upregulated by JAZF1-SUZ12 (red) versus all genes (gray).
(D) Genes (n = 453) significantly upregulated during decidualization of hEnSCs expressing GFP. Left: log2 fold change in expression of these genes in hEnSCs containing GFP or JAZF1-SUZ12 at days 0 and 8 relative to the average expression in hEnSCs containing GFP at day 0. Right: log2 fold change in expression of the same genes in hEnSCs containing JAZF1-SUZ12 at day 8 relative to hEnSCs containing GFP at day 8.
(E) Enrichment (−log10 p value) of representative GO terms in sets of genes upregulated during decidualization of cells containing GFP, upregulated by JAZF1-SUZ12 in non-decidualized cells (day 0), downregulated by JAZF1-SUZ12 in decidualized cells (day 8), or a control set of genes that do not change in expression.
(F) Top: expression of genes (n = 310) previously identified as upregulated in LG-ESS in hEnSCs containing GFP or JAZF1-SUZ12 relative to the average expression in hEnSCs containing GFP. Bottom: gene set enrichment analysis (GSEA) plotting the position of genes upregulated in LG-ESS in the set of genes ranked by change in expression in cells containing JAZF1-SUZ12 versus GFP. NES, normalized enrichment score.
(G) Model. Loss of the SUZ12 N terminus prevents interaction of JAZF1-SUZ12 with EPOP, PALI1, and JARID2, decreasing PRC2 occupancy and reducing H3K27me3 at polycomb target genes. Fusion to JAZF1 induces association with NuA4/TIP60 and increases H4Kac at these sites. These changes are mirrored by ectopic activation of polycomb target genes in hEnSCs, inducing part of the decidualization gene expression program that lacks activation of immune response genes associated with clearance of senescent decidual cells from the endometrium.