Figure 2.
Cell-autonomous changes in neuronal activity regulate the survival of Htr3a+ interneurons
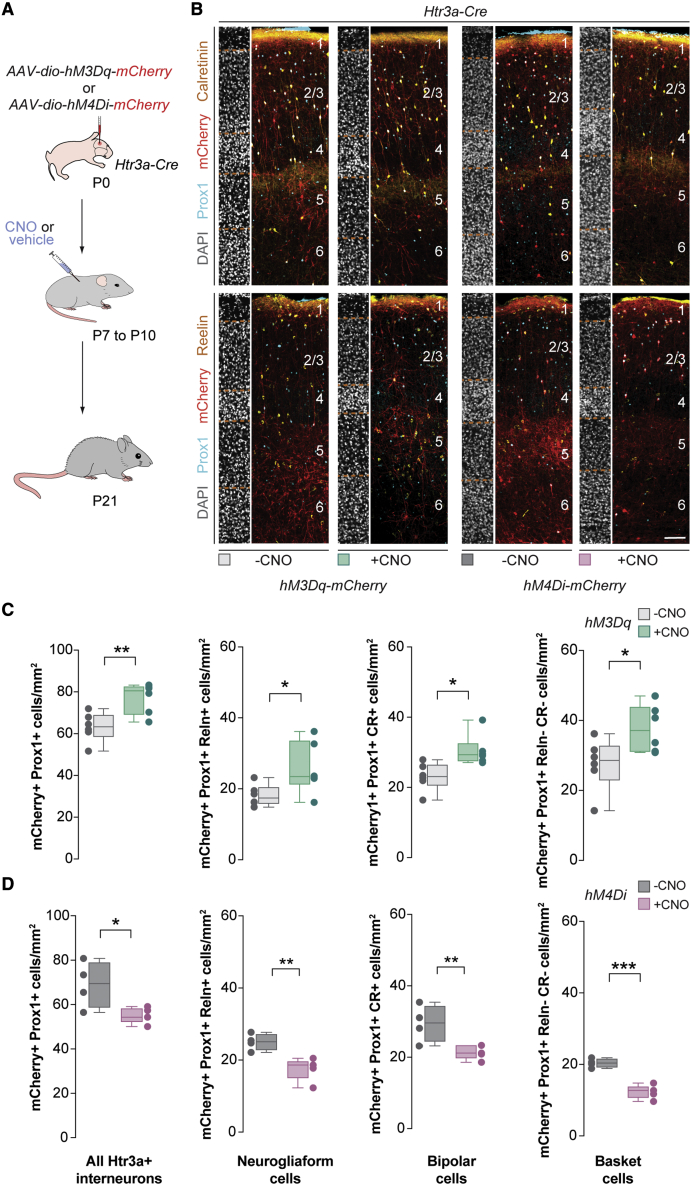
(A) Schematic of experimental design.
(B) Coronal sections through the primary somatosensory cortex of Htr3a-Cre mice at P21 injected with hM3Dq-mCherry (left) or hM4Di-mCherry (right) virus followed by vehicle or CNO treatment immunostained for Prox1 (cyan), mCherry (red), calretinin (yellow, top), and reelin (yellow, bottom). DAPI is shown for counterstaining (gray).
(C) Quantification of the density of all transfected Htr3a+ interneurons (mCherry+, Prox1+), neurogliaform cells (mCherry+, Prox1+, and Reln+), bipolar cells (mCherry+, Prox1+, and CR+), and basket cells (mCherry+, Prox1+, Reln−, and CR−) in control (gray boxplots, n = 6 mice) and CNO-treated mice injected with hM3Dq-mCherry (green boxplots, n = 6 mice) at P21. Prox1+: two-tailed unpaired Student’s t test, ∗∗p = 0.007. Prox1+ Reln+: two-tailed unpaired Student’s t test, ∗p = 0.04. Prox1+ and CR+: two-tailed unpaired Student’s t test, ∗p = 0.01. Prox1+, Reln−, and CR−: two-tailed unpaired Student’s t test, ∗p = 0.03.
(D) Quantification of the density of all transfected Htr3a+ interneurons (mCherry+, Prox1+), neurogliaform cells (mCherry+, Prox1+, and Reln+), bipolar cells (mCherry+, Prox1+, and CR+), and basket cells (mCherry+, Prox1+, Reln−, and CR−) in control (gray boxplots, n = 4 mice) and CNO-treated mice injected with hM4Di-mCherry (red boxplots, n = 5 mice) at P21. Prox1+: two-tailed unpaired Student’s t test, ∗p = 0.03. Prox1+ Reln+: two-tailed unpaired Student’s t test, ∗∗p = 0.006. Prox1+ and CR+: two-tailed unpaired Student’s t test, ∗∗p = 0.005. Prox1+, Reln−, and CR−: two-tailed unpaired Student’s t test, ∗∗∗p = 0.0001.
Data in (C) and (D) are shown as boxplots (median, middle dash), lower and upper quartiles (box borders), and minimum and maximum (whiskers), and the adjacent data points indicate the average cell density in each animal. Scale bar, 100 μm. See also Figure S1.