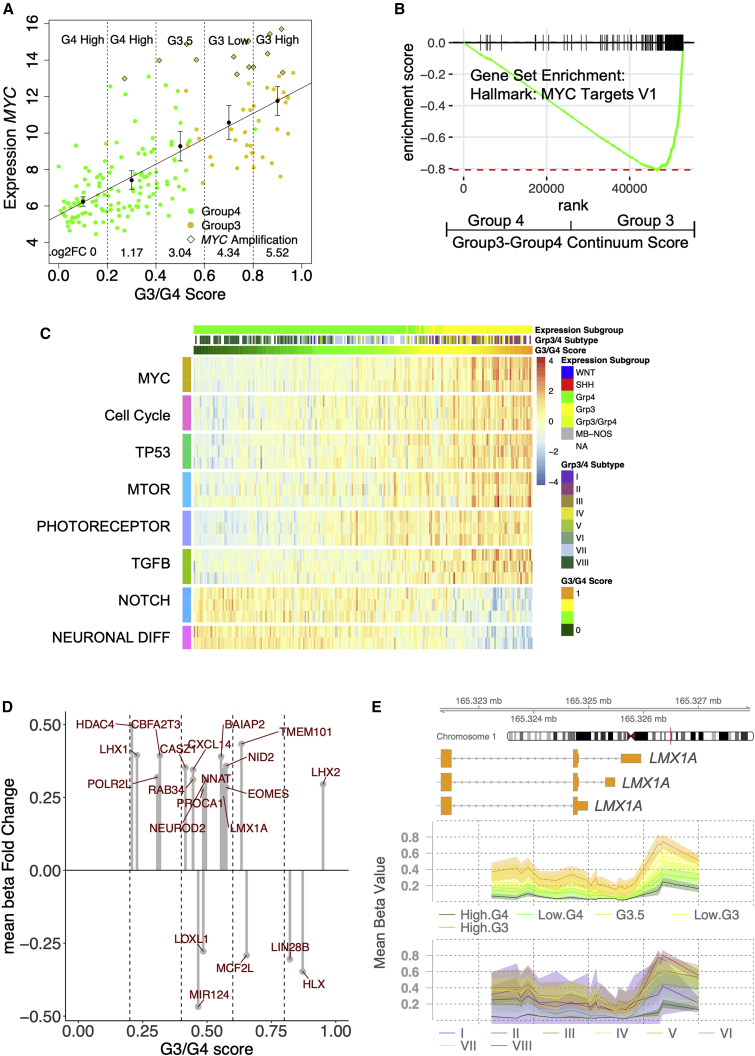
Figure 3.
Position on the group 3/group 4 continuum corresponds linearly to oncogenic pathway activation and methylation of lineage-specific enhancers
(A) Scatterplot showing significant correlation (p < 0.001) between MYC expression and G3/G4 score. Log-linear line of best fit is shown. Dotted lines divide into highG4, lowG4, G3.5, lowG3, and highG3 (these categories are arbitrary divisions of the continuum for the purposes of visualization and comparison and do not represent “real” subgroups), and log2 fold changes for each category relative to highG4 are shown. Error bars represent standard error of mean.
(B) GSEA enrichment plot showing significant enrichment of MYC target genes. Genes were ranked by correlation with G3/G4 score.
(C) Heatmap of ssGSEA results showing level of pathway enrichment for 223 MBGrp3/MBGrp4 individuals ordered by G3/G4 score. MsigDB pathways are curated into pathways (see STAR Methods).
(D) Lollipop plot showing mean beta fold change for DMRs within MBGrp3/MBGrp4 specific enhancers/super-enhancers. The position on the x axis reflects the average point on the continuum at which the methylation level switches from hypo- to hypermethylation.
(E) Plot showing an MBGrp3/MBGrp4-specific enhancer within the MBGrp3-specific gene, LMX1A, which overlaps with a differentially methylated region significantly associated with the G3/G4 continuum. The mean beta value per G3/G4 category (highG4, lowG4, G3.5, lowG3, highG3) and MBGrp3/MBGrp4 subtype (I–VIII) are shown by line and the 95% confidence interval (CI) by shaded area.