Figure 6.
The group 3/group 4 continuum is mirrored in early human cerebellar development
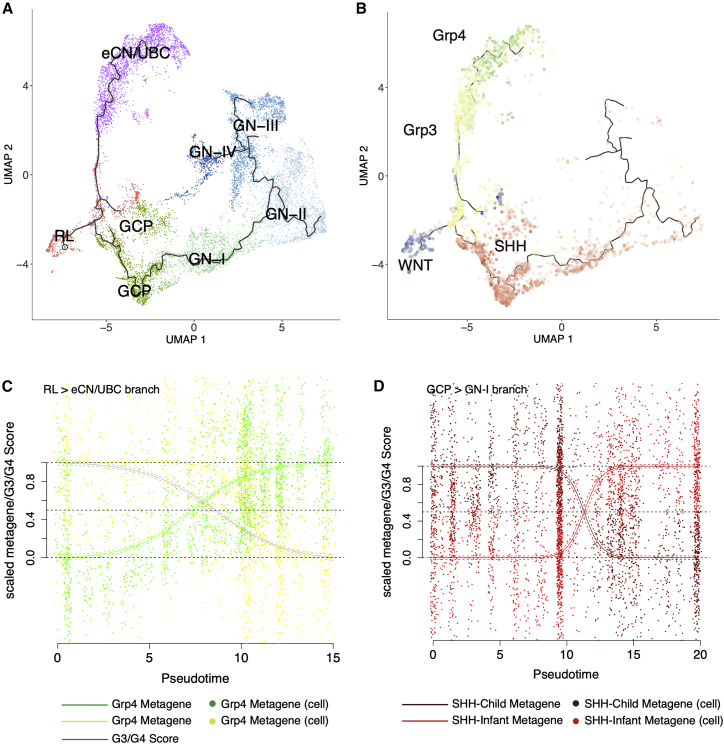
(A) Uniform manifold approximation and projection (UMAP) plot of scRNA-seq profiles showing 12,243 cells of the RL lineage arranged according to developmental trajectory, which is indicated by the black line. Color denotes cell type as determined by graph-based clustering; RL, rhombic lip precursors; GCP, granule cell precursors; GN-I, GN-II, GN-III, GN-IV, 4 granule neuron cell types; eCN/UBC, excitatory cerebellar neurons/unipolar brush cells.
(B) UMAP plot of the RL lineage with those cells within the top decile of metagene expression marked with the following colors: MBGrp4, green; MBGrp3, yellow; MBSHH, red; MBWNT, blue.
(C) Scatterplot showing per-cell scaled metagene expression along the RL to eCN/UBC branch. Fitted sigmoid curves are shown, with SD indicated as dashed lines. The gray line represents a sigmoid curve fitted to per-cell G3/G4 score as a function of pseudotime.
(D) Scatterplot showing per-cell scaled metagene expression along the GCP to GN branch. Fitted curves are shown with SD shown as dashed lines. Curves are scaled to be constrained to a range of 0 and 1, to be coherent with bulk analysis. For this reason, by definition, some individual cells lie outside the 0 and 1 range.