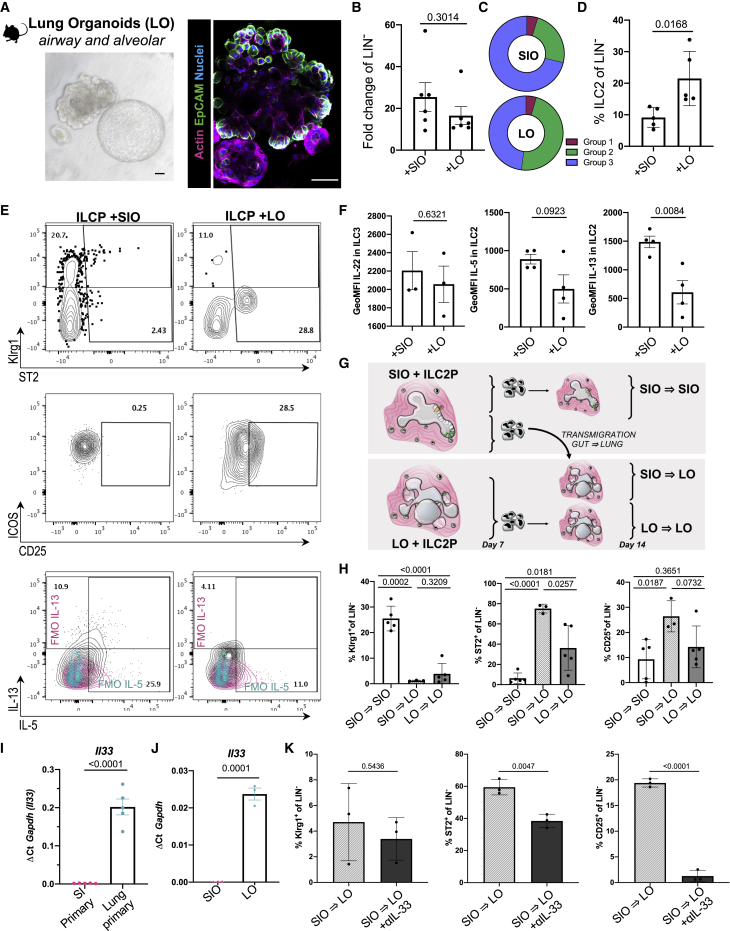
Figure 4.
Co-culture with gut and lung organoids (LOs) drive tissue-specific ILC2 imprinting
(A) Representative bright-field and confocal image of murine primary distal lung epithelial organoid showing cystic or saccular structures (scale bars: 25 μm).
(B) Fold change expansion of Live, EpCAM−, CD45+ ILCPs after 7-day co-culture with SIOs or LOs (N = 6 animals, two independent experiments).
(C) Relative proportion of mature ILC groups, showing Live, EpCAM−, CD45+ group 1 ILCs (magenta, Live EpCAM−CD45+Lineage−RORγt−NKp46+NK1.1+Gata3−), ILC2 (green, gated as Live EpCAM−CD45+Lineage−RORγt−NKp46−NK1.1−Gata3+), and ILC3 (blue, EpCAM−CD45+Lineage−RORγt+NKp46+/−NK1.1+/−Gata3−) derived from 7-day co-culture of ILCPs with SIOs or LOs.
(D) Quantification of absolute ILC2 frequency in (C) as a proportion of all LIN− cells (N = 5 animals).
(E) Representative flow plots of Klrg1, ST2, ICOS, CD25, IL-13, and IL-5 expression in EpCAM−CD45+NKp46−NK1.1−RORγt− putative ILC2s after co-culture with SIOs or with LOs (FMOs, magenta and blue), with the GeoMFI of IL-5 and IL-13 cytokine expression in ILC2 and IL-22 expression in RORγt+ ILC3 quantified in (F) after 4-h unbiased PMA/Ionomycin stimulation (N = 4, two experiments).
(G) Schematic of SIO-to-LO experimental design.
(H) Frequency of Klrg1+, ST2+, and CD25+ ILC2s after 14-day co-culture (N = 3–5 animals).
(I) Relative gene expression of Il33 in whole primary small intestine and lung tissue (N = 5 animals), as well as in SIOs and LOs (n = 3 wells of organoids) in (J).
(K) Frequency of Klrg1+, ST2+, and CD25+ ILC2s in SIO-to-LO co-cultures with and without neutralizing dose of rmIL-33 blocking antibody (50 ng/mL, N = 3 animals).
Error bars represent SEM; p values are from unpaired Student’s t tests.