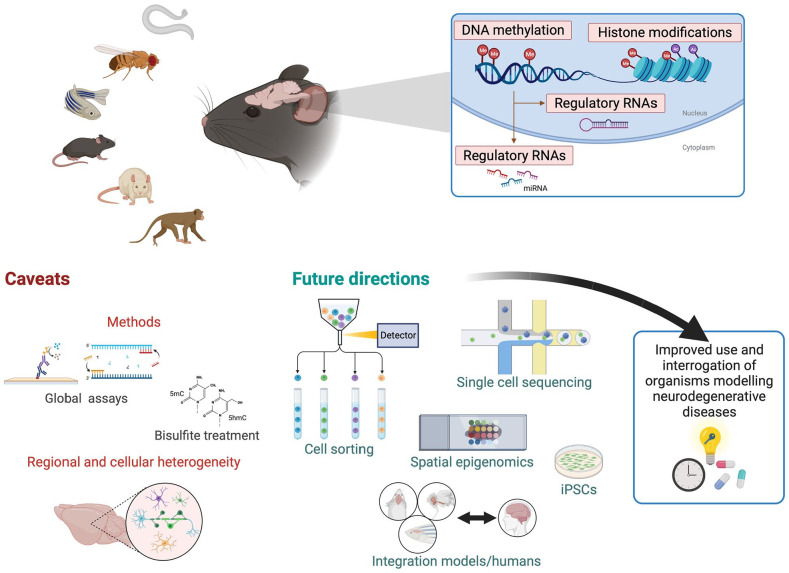
Figure 1.
Vertebrate and invertebrate models useful for epigenetics research of neurodegerative diseases, epigenetic marks investigated to date, common caveats, and future directions to improve the use and benefits of these powerful models. Model organisms used to study neuroepigenetics of neurodegenerative diseases (top left) range from simpler organisms such as nematodes (Caenorhabditis elegans), fruit fly (Drosophila melanogaster) and zebra fish (Danio rerio), rodents (mice and rats), and non-human primates. DNA methylation, histone modifications and regulatory RNAs (predominantly miRNAs) comprise epigenetic processes that have been studied in these models (top right). Caveats of studies to date (bottom left) include methods commonly employed, many assessing global levels of epigenetic modifications (global assays), and—in the case of DNA methylation—the vast majority relying in bisulfite conversion, which does not allow to differentiate between 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC). Of great importance in epigenetics research, regional and cellular heterogeneity have predominantly not been taken into account and/or explored. Future research (bottom right) must mitigate limitations of studies thus far by discerning tissue- and cell-specific signatures, take advantage of immerging single-cell and spatial technologies, integrate findings from model organisms with human studies, and consider additional strategies such as the parallel use of iPSCs. Taken together with the development of better disease models currently underway, future research should aim to improve the use of model organisms for the understanding of epigenetic processes in neurogenerative conditions, and of how and when we should modify and manipulate aspects of human disease, particularly important for drug discovery and testing.
Created with BioRender.com.