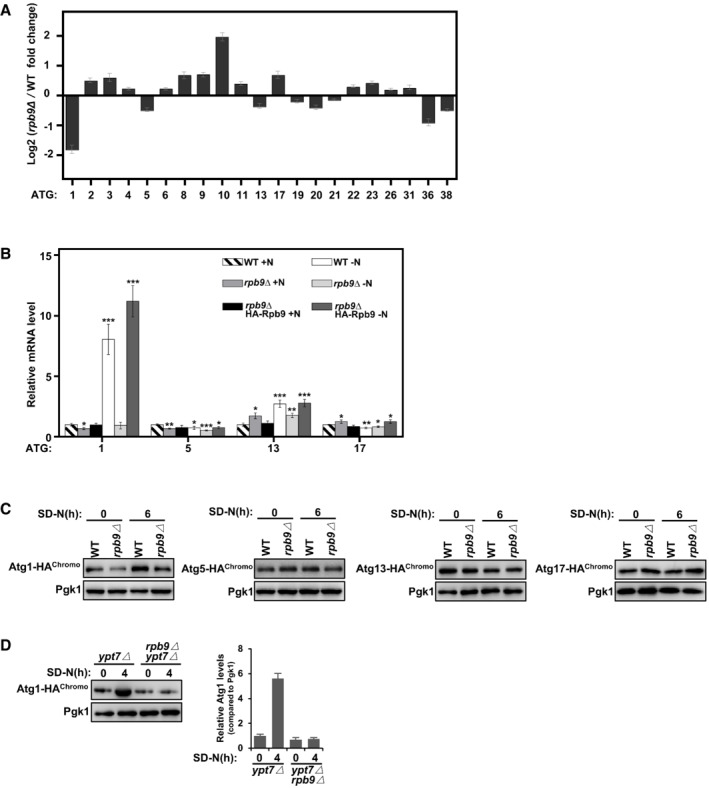
Figure EV3. Rpb9 is important for ATG1 transcription.
-
AData from genome‐wide analysis of differential gene expression in rpb9∆ yeast cells compared to WT cells showed Rpb9 deletion caused specific downregulation of the ATG1 gene. Bars represent mean, error bars represent standard deviation (n = 5 biological replicates).
-
BWT, rpb9∆, and rpb9type="InMathematical_Operators">∆+HA‐Rpb9 yeast cells were grown to log phase in YPD (+N) and then shifted to nitrogen starvation (−N) for 3 h. mRNA levels of ATG1, ATG5, ATG13, and ATG17 were quantified by qRT‐PCR. Bars represent mean, error bars represent standard deviation, significance was determined by one‐way ANOVA (unpaired) followed by Tukey's multiple comparison test, *indicates P < 0.05, **indicates P < 0.01, ***indicates P < 0.001 (n = 5 biological replicates).
-
CProtein expression levels of chromosome tagged Atg1, Atg5, Atg13, and Atg17 in WT and rpb9∆ yeast cells were analyzed before and after SD‐N starvation.
-
DAtg1 was HA‐tagged at the C‐terminal by chromosome recombination in ypt7∆ cells and in ypt7∆ rpb9∆ cells and its protein levels were analyzed before and after nitrogen starvation in an SD‐N medium.
