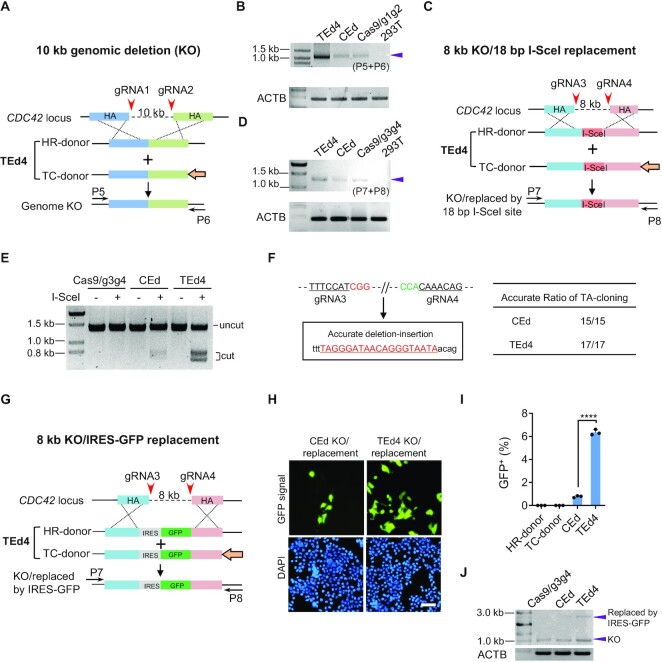
Figure 5.
TEd-mediated long genomic deletion and replacement in HEK293T cells. (A) Schematic presentation of a 10 kb genome knock-out (KO) at the CDC42 locus by TEd4. Paired gRNAs spaced ∼10 kb (gRNA1 + gRNA2) apart were designed to target the CDC42 locus. Primers used to amplify the target genomic deletion are marked as (P5 + P6). (B) Representative PCR amplicons of the junction region using the primers indicated in (A). HEK293T cells served as a negative control. The representative amplicon of the targeted deletion is marked with an arrow. (C) Schematic presentation of an 8 kb genome KO at the CDC42 locus and simultaneous replacement with an I-SceI recognition site (18 bp) at the target site by TEd4. Paired gRNAs spaced ∼8 kb (gRNA3 + gRNA4) apart were designed as indicated to target the CDC42 locus. Primers used to amplify the target genomic regions are marked as (P7 + P8). (D) Representative PCRs in the junction region using the primers indicated in (C). HEK293T cells served as a negative control. The representative amplicon of the targeted deletion is marked with an arrow. (E) Amplicons from CEd and TEd were incubated with or without I-SceI enzyme and analyzed on an agarose gel. Digested products are marked as ‘cut’ while the original amplicon is marked as ‘uncut’. (F) Evaluation of accurate replacement of 18 bp I-SceI recognition sequence in CEd and TEd4 by TA cloning sequencing (CEd, 15/15; TEd, 17/17). Two PAM sequences of gRNAs are shown in red and green. The 18 bp intended insertion is underlined. (G) Schematic presentation of an 8 kb genome KO at the CDC42 locus and simultaneous replacement with IRES–GFP (∼1.2 kb) at the target site by TEd4. (H) Living cell fluorescence images of the GFP pan-cellular signal of CEd and TEd4 in (G) observed at 3 days after transfection (unsorted). Scale bar, 50 μm. (I) Efficiencies of IRES–GFP replacement in (G) were evaluated by FACS. (J) Representative PCR amplicons of the target region using the primers indicated in (G). The deletion amplicons are marked with two bands in CEd and TEd4. The lower bands are the expected KO sizes, the upper bands of CEd and TEd4 are marked with their expected sizes after IRES–GFP insertion (upper arrow). All data are presented with individual data points and mean ± SD for n = 3 independent biological replicates. P-values were obtained using the two-tailed Student's t-test. ***P< 0.001, ****P< 0.0001.