Figure 1.
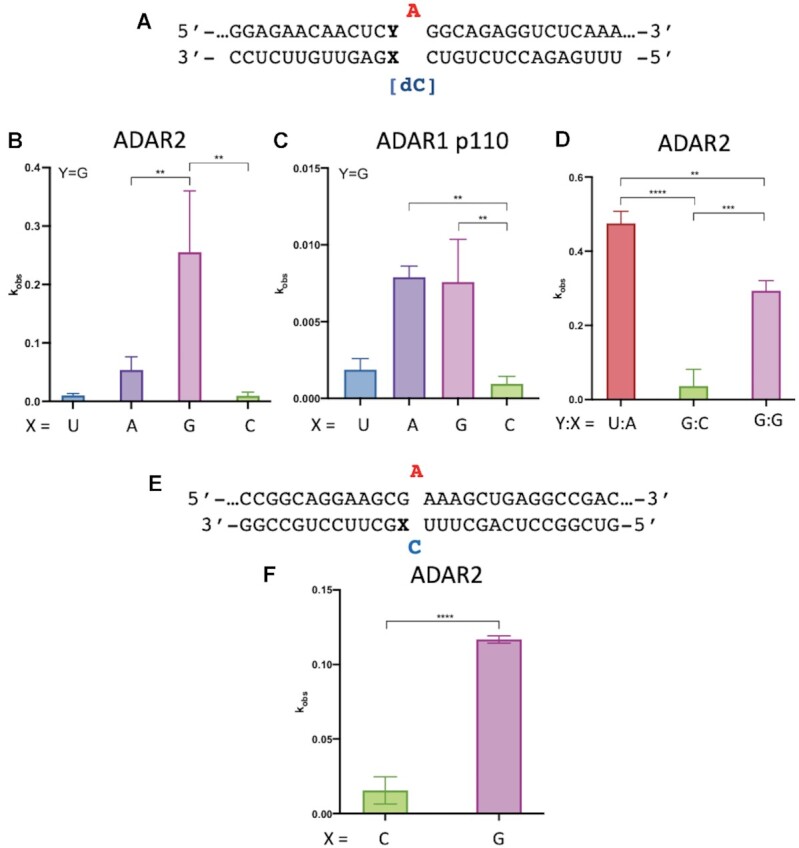
In vitro deamination kinetics for ADAR2 and ADAR1 p110 varying 5′ nearest neighbor base pairing. (A) Sequence of model substrate for ADARs with varying base pairing adjacent to the editing site (X:Y). The target sequences are derived from the human IDUA mRNA. See Table 1 for fitted values. (B) Comparison of rate constants for reaction with 100 nM ADAR2. (C) Comparison of rate constants with 250 nM ADAR1 p110. (D) Comparison of rate constants with 10 nM ADAR2. (E) Duplex substrates where target sequence is derived from wild type human MECP2 mRNA varying base pairing with 5′ G adjacent to the editing site (X). (F) Comparison of rate constants for reaction with 100 nM ADAR2. See Table 1 for fitted values. Plotted values are the means of three technical replicates ± standard deviation. Statistical significance between groups was determined using one-way ANOVA with Tukey's multiple comparisons test or an unpaired t-test with Welch's correction; **P < 0.01; ***P < 0.001; ****P < 0.0001.
