Figure 1.
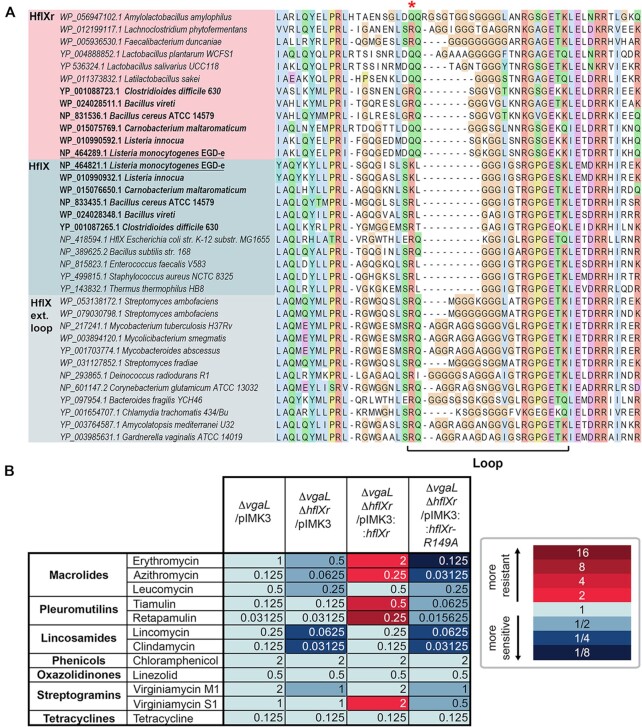
Sequence alignments of HflX and HflXr proteins and MIC data. (A) Sequence alignment of the resistance-associated loop region within the N-terminal domain of selected HflX (blue) and HflXr (pink) representatives, showing independently evolved insertions in HflXr and HflX. Taxa in bold are those with both HflX and HflXr. Conserved R/Q residue (R149 in L. monocytogenes HflXr) is marked with a red asterisk. The full alignment is found in Supplementary Data S1. (B) Minimum inhibitory concentrations (MICs) of ribosome-targeting antibiotics against L. monocytogenes EGDe strains lacking or expressing HflXr or/and VgaL/Lmo0919 ARE-ABCF. The color code is made with respect to the first column that contains the ΔvgaL/pIMK3 MICs.