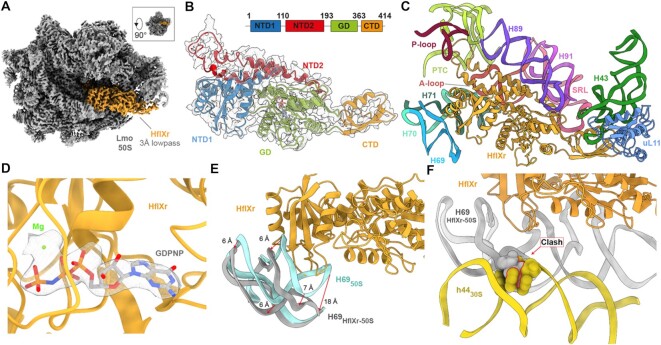
Figure 2.
Cryo-EM structure of the L. monocytogenes HflXr-50S complex. (A) Cryo-EM density (3 Å low-pass filtered) of the 50S ribosomal subunit (grey) with HflXr (orange). Inset shows relative orientation of (A) to crown view. (B) Isolated cryo-EM map density (grey mesh; 3 Å lowpass filtered) with a molecular model for HflXr colored according to domains: N-terminal subdomain I (NTD1, blue), N-terminal subdomain II (NTD2, red), G-domain (GD, green) and C-terminal domain (CTD, orange). (C) Interactions of HflXr (orange) NTD1 with H69 (blue), H70 (cyan), H71 (dark green), the α-helices of NTD2 with H89 (purple) and H91 (light purple), NTD2-loop with the PTC (lime) and the CTD with uL11 (light blue). (D) GDPNP (grey) in extracted density (grey mesh) within the binding pocket of the GD (orange) with a putative coordinated magnesium ion (green). (E) Binding of HflXr (orange) causes a shift in H69 (grey) compared to the vacant L. monocytogenes ribosome (light blue, aligned on 23S rRNA). (F) H69 (grey) movement caused by HflXr would sterically clash with h44 of L. monocytogenes 30S subunit (yellow, PDBID: 7NHN; (11)) on the 70S ribosome, when aligned on the basis of 23S rRNA.