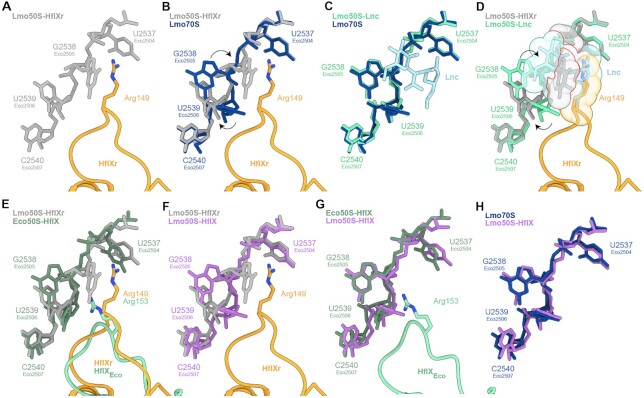
Figure 5.
HflXr-induced conformational changes at the PTC are incompatible with lincomycin binding. (A) NTD2-loop Arg149 of HflXr (orange) with selected L. monocytogenes 23S rRNA nucleotides (grey). (B) as (A), but superimposed with L. monocytogenes 23S rRNA nucleotides (dark blue) from vacant 70S ribosome. Nucleotide rearrangements induced by HflXr are indicated with black arrows. (C) Comparison of L. monocytogenes 23S rRNA nucleotides in presence (green) and absence (dark blue) of lincomycin (Lnc, light blue). (D) as (A), but superimposed with Lnc (light blue) bound to the L. monocytogenes 23S rRNA nucleotides (cyan) with nucleotide rearrangements induced by HflXr indicated with black arrows and shown as spheres with red lines indicating steric clashes. (E, F) as (A), but superimposed with (E) E. coli HflX-50S complex (PDB ID 5ADY) (17), and (F) 23S rRNA nucleotides (purple) from the L. monocytogenes 70S refined into the L. monocytogenes HflX-50S complex. (G, H) Comparison of 23S rRNA nucleotides from L. monocytogenes HflX-50S complex with (G) E. coli HflX-50S complex (PDB ID 5ADY) (17), and (H) L. monocytogenes 70S ribosome.