Fig. 1. Cryo-EM structure of huPrP23–144 amyloid fibrils.
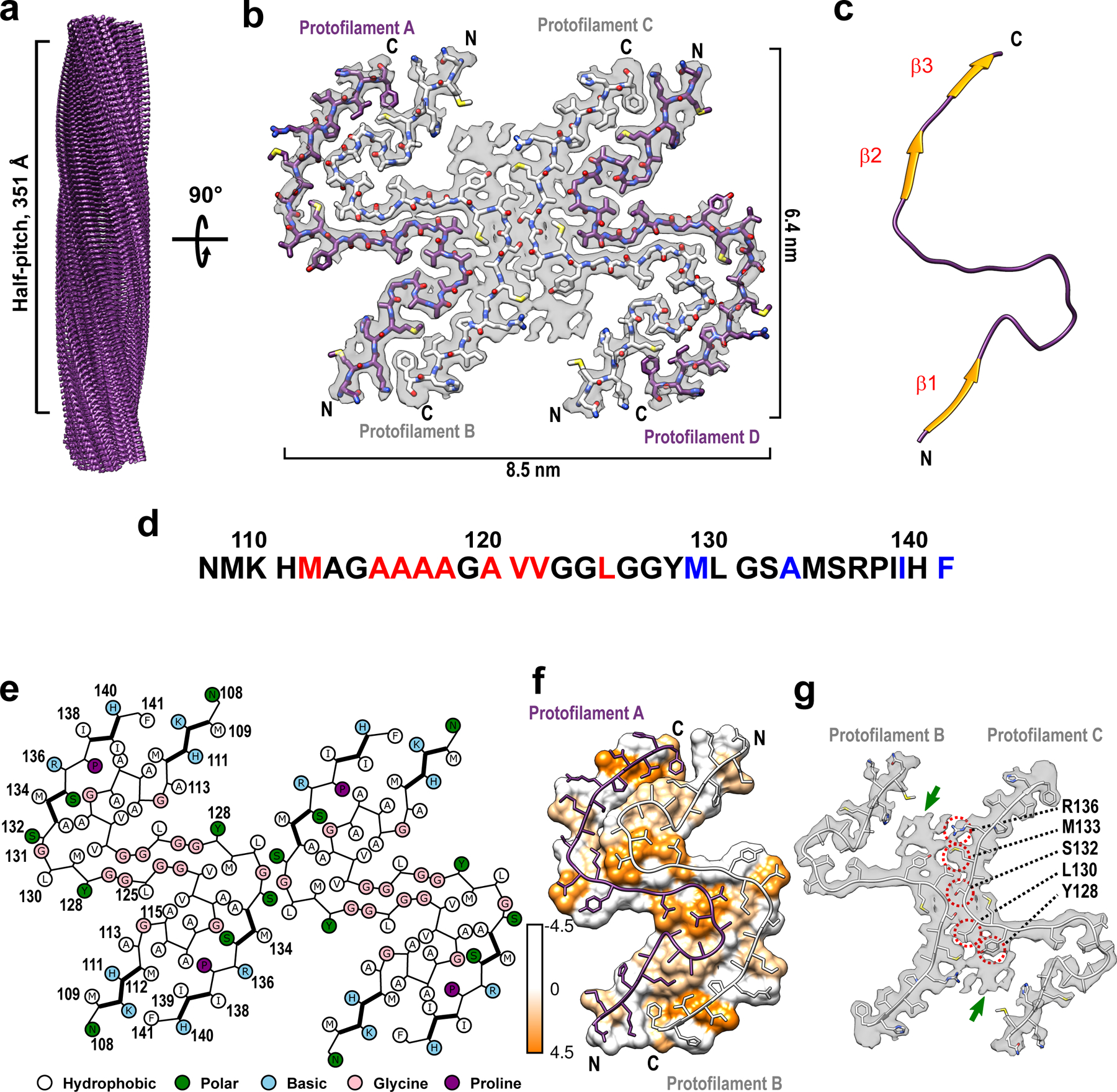
a Cryo-EM density map showing a left-handed twisted helix with a half-pitch of 351 Å and a classical parallel, in-register β-sheet architecture. b The top view of the atomic model superimposed on the cryo-EM density map. The amyloid core contains four identical protofilaments. Two outer protofilaments with greater accessibility to water are depicted in purple; two inner protofilaments are depicted in white. c One representative subunit of the fibril core with β-strands shown in orange. d Amino acid sequence for the highly ordered core of huPrP23–144 fibrils. N- and C-terminal solvent-inaccessible hydrophobic residues are shown in red and blue, respectively. e Schematic representation of one cross-sectional layer of the amyloid core, with β-strands shown as thicker lines. f Hydrophobicity of the cross-section of protofilaments A and B, with hydrophobicity levels colored according to Kyte-Doolittle21 (top view). g One cross-sectional layer of protofilaments B and C shown as atomic model superimposed on the cryo-EM map (top view). Extra unassigned densities (likely representing side chains of residues outside the core region) are indicated by green arrows; residues within the amyloid core involved in the inter-protofilament interactions are indicated by red circles.
