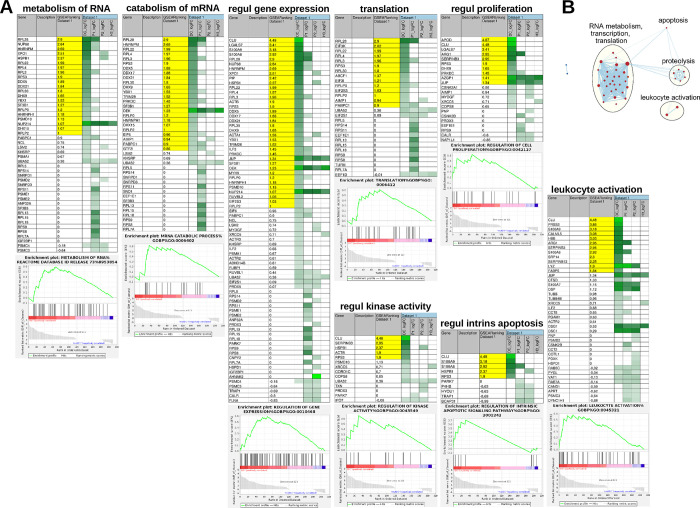
Fig 6. Bioinformatic analysis of biological processes comparing the interactome of DEK/NUP214 with those of its mutants by GSEA and Cytoscape–quantification of interactions.
The filter in GSEA was set at a minimum of 10 genes/set. A. Shown are the eight most statistically significant processes revealed by GSEA. DC_logFC, P1_logFC, P2_logFC, and H3_logFC- the data set names given by GSEA to the list of interactors of DEK/NUP214, ΔP1/NUP214, ΔP2/NUP214, and DEK/ΔH3B, respectively. Yellow—leading-edge subset proteins responsible for the enrichment of the respective gene sets. The intensities of green correlate with the strength of interaction between DEK/NUP214, ΔP1/NUP214, ΔP2/NUP214, or DEK/ΔH3B and the given interactors. B. The analysis of the interactomes of DEK/NUP214 and its mutants in GSEA was refined in Cytoscape/EnrichmentMap, showing the processes highly enriched in the interactome of DEK/NUP214.