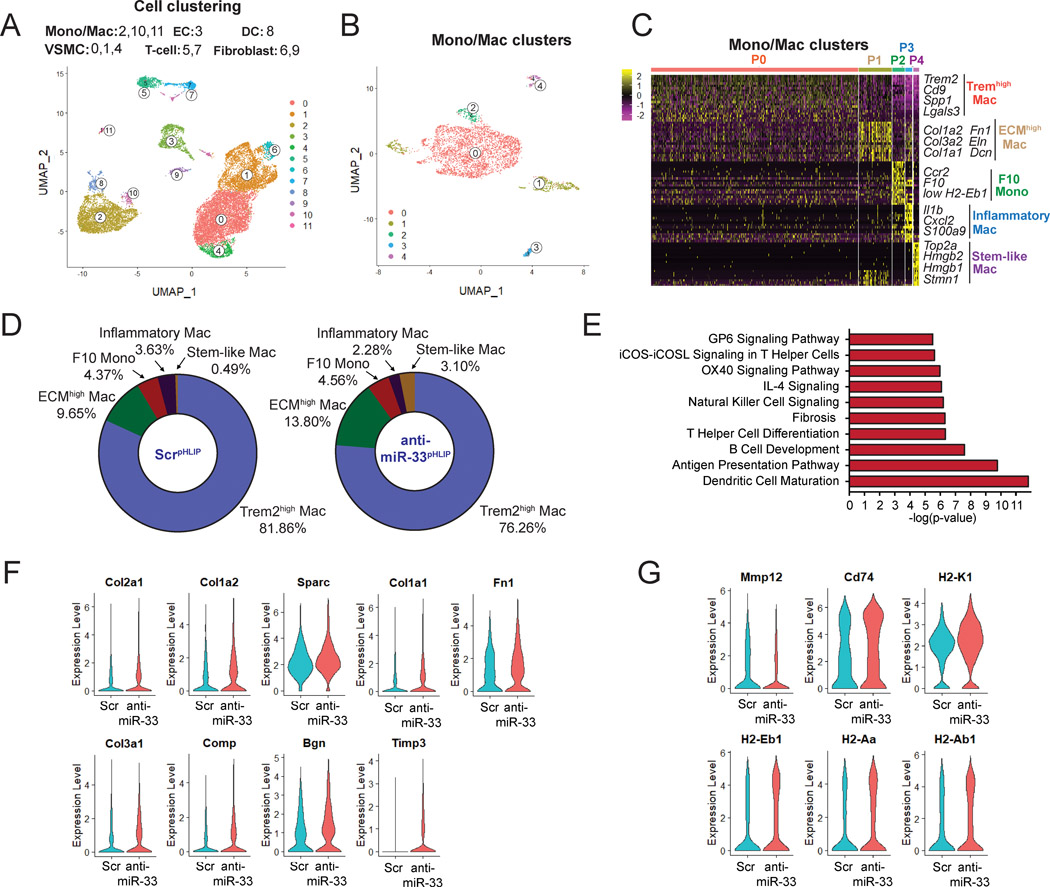
Figure 5. Single cell RNA-Seq analysis reveals increased expression of extracellular matrix (ECM) genes in macrophages isolated from anti-miR-33pHLIP treated mice.
A, Uniform manifold approximation and projection (UMAP) representation of aligned gene expression data in single cells extracted from atherosclerotic plaques of anti-miR-33pHLIP and ScrpHLIP-treated mice (n=3 per group). UMAP dimensionality reduction analysis identified 12 major clusters. B, UMAP representation of aligned gene expression data in monocytes and macrophages (Mono/Mac) cluster extracted from atherosclerotic plaques. C, Heatmap of the 20 most upregulated genes in each cluster defined in B and selected enriched genes used for biological identification of each cluster. D, Proportions of defined Mono/Mac populations extracted from the atherosclerotic aortas of anti-miR-33pHLIP and ScrpHLIP-treated mice. E, Pathway enrichment of differentially expressed genes expressed as the log[–P] analyzed by Ingenuity Pathway Analysis. F, Violin plots of the top differentially expressed genes showing statistically significant upregulation of ECM in Mono/Mac population from anti-miR-33pHLIP-treated mice. G, Violin plots of the top differentially expressed genes showing less inflammatory and higher antigen presentation genes in Mono/Mac population from anti-miR-33pHLIP-treated mice.