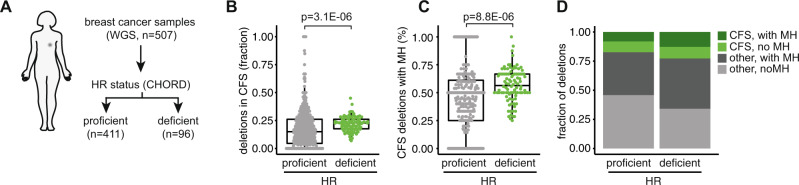
Fig. 6. Analysis of genomic deletions at common fragile sites.
A Whole-genome sequence (WGS) data of n = 507 breast cancers of the International Cancer Genome Consortium (ICGC) were analyzed, of which n = 96 of which were HRD. HRD status was assessed using the CHORD algorithm. B, C Allelic deletions from 507 breast cancers mapping to common fragile sites were plotted in panel B. Allelic deletions mapping to common fragile sites and having microhomology (MH) (larger or equal to 2 bp) at the breakpoints were plotted in panel C. Box plots depict the mean (center line), 25th and 75th percentiles (box boundaries), and the largest values no more than 1.5* the interquartile range (whiskers). Statistical analysis was done using an FDR-corrected two-sided Wilcox test. D Analysis of deletions within CFS with or without MH as a percentage of total deletions in 507 breast cancers were plotted.