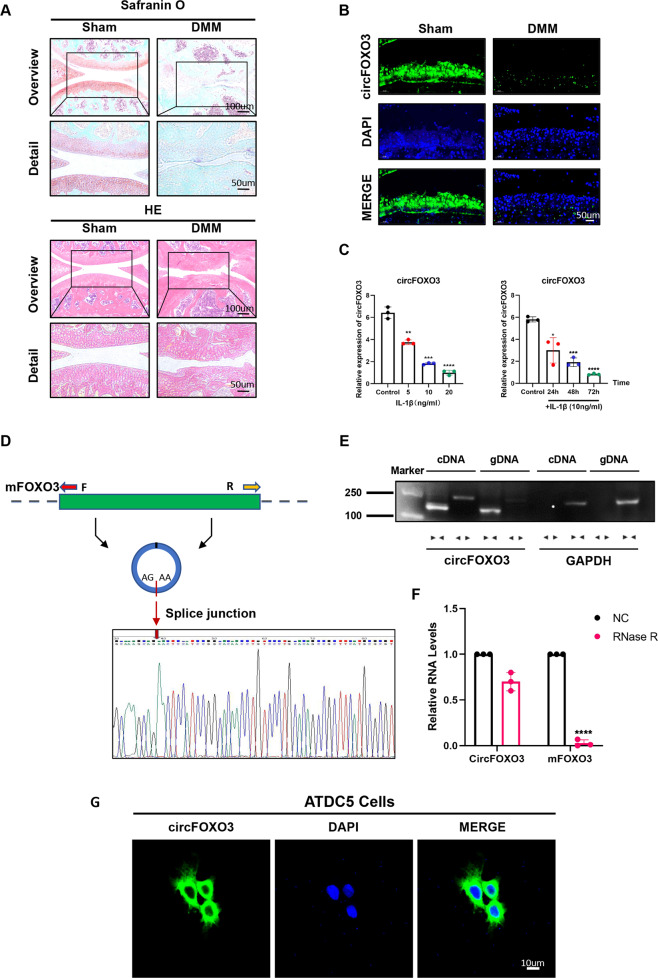
Fig. 1. CircFOXO3 exhibits lower expression in OA tissue and predominantly localized in the cytoplasm.
A Safranin-O/fast green staining and H&E staining of the cartilage from different groups. Scale bars, 100 µm and 50 µm. B Expression levels of circFOXO3 was detected by RNA FISH in C57BL/6 samples. Representative photomicrographs and fluorescence intensity of FISH are shown. Scale bar, 50 µm. C Expression of circFOXO3 after treating with IL-1β for different concentrations and time spans. (n = 3) *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. D Sanger sequencing after PCR using the indicated divergent flanking primers confirmed the “head-to-tail” splicing of circFOXO3. E The presence of circFOXO3 was validated in ATDC5s, circFOXO3 was amplified by divergent primers in cDNA but not gDNA. GAPDH was used as a negative control. F After treating or without treating with RNase R, the expression of circFOXO3 and FOXO3 mRNA in ATDC5s were detected by RT-qPCR. (n = 3) ****p < 0.0001. G RNA FISH showed that circFOXO3 was predominantly localized in the cytoplasm. CircFOXO3 probes were labeled with Cy-3. The nuclei were stained with DAPI. Scale bar, 10 µm.