Figure 5.
Variability in the firing properties of cerebellar nuclei neurons and Purkinje cells in mouse models of disease
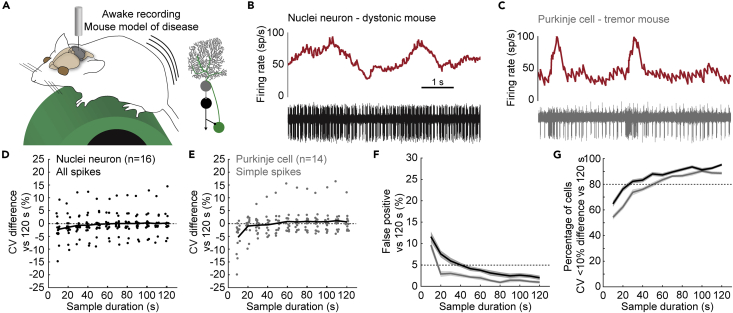
(A) Schematic of recording setup depicting an awake mouse on a foam running wheel (left). Simplified schematic of cerebellar circuit (right). Neurons included in the analysis for this figure come from Ptf1aCre;VGlut2fl/fl (dystonia-like) mice8; harmaline-injected (tremor) mice7; and L7Cre;Vgatfl/fl (ataxia) mice.7
(B) Example trace of a nuclei neuron recording in an awake dystonic mouse. Firing rate as calculated over 0.5 s intervals (top) and matched raw electrophysiological recording (bottom).
(C) Example trace of a Purkinje cell recording in an awake tremoring mouse. Firing rate as calculated over 0.5 s intervals (top) and matched raw electrophysiological recording (bottom).
(D and E) Mean difference in (D) nuclei neuron spike, and (E) Purkinje cell simple spike CV estimates between 100 sample durations and one representative reference duration. Each dot represents the average for 1 cell. Black line represents the mean for all cells included in the analyses.
(F) Number of significant paired t-tests between CV estimate in 120 s-long reference duration and 100 samples for each sample duration.
(G) Percentage of CV estimates in 100 samples for each sample duration within 10% deviation of the reference duration. For F and G: Mean of 25 sets of 1 reference duration with 100 sample duration each = solid line; ± SEM = shaded region. Nuclei neuron all spikes in green; Purkinje cell simple spikes in gray.