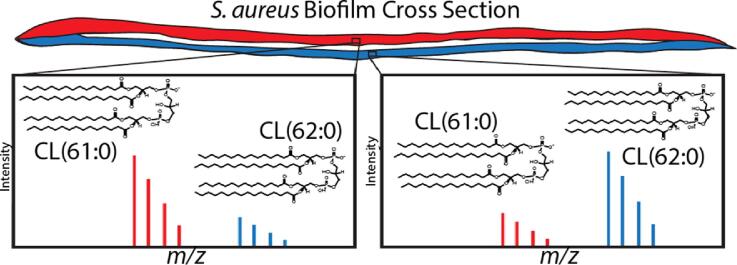
Graphical abstract
Three distinct horizontal layers are visible within the cross section of an S. aureus biofilm where some cardiolipin species such as CL(61:0) are more abundant in the top layer (red), and other species such as CL(62:0) are more abundant in the bottom layer (blue).
Keywords: Imaging Mass Spectrometry, Trapped Ion Mobility, Biofilms, Lipids
Abbreviations: CASI, continuous accumulation of selected ions; CMC, carboxymethylcellulose; CL, Cardiolipin; CFU, colony forming units; DAN, 1′,5′-Diaminonaphthalene; DAPI, 4′,6-diamidino-2-phenylindole; DG, diacylglycerol; DGDG, digalactosyldiacylglycerol; DHA, 2′,5′-Dihydroxyacetophenone; DsRed, red fluorescent protein from Discosoma; EFG, electric field gradient; FWHM, full-width half max; GFP, green fluorescent protein; IMS, imaging mass spectrometry; ITO, indium-tin oxide; L-PG, lysyl-phosphatidylglycerol; MALDI, matrix-assisted laser desorption/ionization; OCT, optimal cutting temperature; PG, phosphatidylglycerol; PIP, phosphatidylinositol phosphate; PC, phosphatidylcholine; S/N, Signal-to-noise; TIMS, trapped ion mobility spectrometry; TSA, tryptic soy agar; TSB, tryptic soy broth; TIC, total ion current; XIM, extracted ion mobilogram
Highlights
-
•
Spatially resolved lipidomic analysis of horizontal S. aureus biofilm layers shows that they are molecularly distinct.
-
•
Implementation of Trapped Ion Mobility Spectrometry increases sensitivity and peak capacity.
-
•
S. aureus biofilms grown anaerobically do not form horizontal layers, and have homogenous lipid distributions.
Abstract
Introduction
Although Staphylococcus aureus is the leading cause of biofilm-related infections, the lipidomic distributions within these biofilms is poorly understood. Here, lipidomic mapping of S. aureus biofilm cross-sections was performed to investigate heterogeneity between horizontal biofilm layers.
Methods
S. aureus biofilms were grown statically, embedded in a mixture of carboxymethylcellulose/gelatin, and prepared for downstream matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS). Trapped ion mobility spectrometry (TIMS) was also applied prior to mass analysis.
Results
Implementation of TIMS led to a ∼ threefold increase in the number of lipid species detected. Washing biofilm samples with ammonium formate (150 mM) increased signal intensity for some bacterial lipids by as much as tenfold, with minimal disruption of the biofilm structure. MALDI TIMS IMS revealed that most lipids localize primarily to a single biofilm layer, and species from the same lipid class such as cardiolipins CL(57:0) – CL(66:0) display starkly different localizations, exhibiting between 1.5 and 6.3-fold intensity differences between layers (n = 3, p < 0.03). No horizontal layers were observed within biofilms grown anaerobically, and lipids were distributed homogenously.
Conclusions
High spatial resolution analysis of S. aureus biofilm cross-sections by MALDI TIMS IMS revealed stark lipidomic heterogeneity between horizontal S. aureus biofilm layers demonstrating that each layer was molecularly distinct. Finally, this workflow uncovered an absence of layers in biofilms grown under anaerobic conditions, possibly indicating that oxygen contributes to the observed heterogeneity under aerobic conditions. Future applications of this workflow to study spatially localized molecular responses to antimicrobials could provide new therapeutic strategies.
Introduction
Staphylococcus aureus is a Gram-positive opportunistic pathogen responsible for over 10,000 deaths and 320,000 hospitalizations per year in the United States and is a leading cause of healthcare-associated infections, such as endocarditis and bacteremia [1], [2]. In addition, S. aureus is also the leading pathogen in biofilm-related infections, further confounding its burden on human health [3]. Biofilms are complex bacterial communities that adhere to a surface, such as surgical prosthetics or biological tissue, in infection [4], [5]. Bacteria within these communities display heterogeneous behavior, and S. aureus biofilms can develop spatially discrete horizontal layers comprised of bacteria in different phenotypic states. Biofilm development leads to changes in characteristics compared to planktonic bacteria, including increased stress tolerance and altered gene regulation [6], [7], [8], [9], and can involve changes in the bacterial lipidome [10]. Lipids play important roles in S. aureus ranging from membrane composition to evasion of the host immune response [11], [12], [13]. Bacterial metabolic processes, stress response, and virulence factor production can also be affected by changes in lipid composition [14], [15]. The S. aureus lipidome consists primarily of phospholipids, including phosphatidylglycerols (PG), phosphatidylinositol phosphates (PIP), cardiolipins (CL), and lysyl-phosphatidylglycerols (L-PG) [14], [16], [17], [18], [19], each with distinct roles in bacterial functions. While the functions of these lipids are well described within an S. aureus bacterium, little is known about the roles they may play in larger bacterial colonies, such as an S. aureus biofilm. Determining where bacterial lipids localize within horizontal layers of S. aureus biofilms under basal conditions is necessary to provide insight into how the these bacterial colonies respond to external factors, such as differences in oxygen availability.
Many previous studies have used lipid extracts from cultured bacteria to investigate the lipidome of S. aureus for research interests ranging from membrane rigidity to antibiotic resistance [16], [18], [20], [21]. These approaches provide quantitation of the overall lipid content, but come at the expense of spatial information that is especially important in biofilm formation. Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) [22] is a technology that is effective for mapping hundreds-to-thousands of molecules, including small metabolites [23], [24], glycans [25], [26], [27], lipids [28], [29], peptides [30], [31] and intact proteins [32], [33] in tissue at biologically-relevant spatial resolutions [22], [34]. The ability to interrogate molecular distributions within bacterial communities by MALDI IMS has been previously demonstrated for a wide range of microbial molecules including phenol-soluble modulins [35], [36], [37], metabolites [35], [38], [39], [40], [41], [42], [43], [44], [45], oligosaccharides [46] and some lipids [40], [41], [46], [47], [48]. However, most of this work has been focused on analytes other than lipids, or has been performed with bacteria other than S. aureus or a co-culture of S. aureus with other microbes. Lipidomic investigations of monomicrobial S. aureus biofilms, however, would offer insight to the basal phenotypic state of bacterial lipid distributions. In addition, most prior examples of MALDI IMS of microbial molecules have been performed by sampling the top of the biofilm, investigating molecular distributions on the surface of the biofilm. A few examples of biofilm cross-sections have been demonstrated for the investigation of microbial metabolites within the cross-sectional depth of the biofilm [38], [39], but none have investigated S. aureus lipid distributions within horizontal biofilm layers.
One challenge posed by microbial imaging is the small size of an individual bacterium. Most MALDI IMS studies of bacterial communities have been performed at spatial resolutions greater than 50 µm, with rare exceptions [47], [45]. Because these S. aureus biofilm layers are approximately 100 µm thick, higher spatial resolutions are necessary to effectively investigate inter-layer variations. To this end, we sought to increase the spatial resolution of our lipidomic study (10 µm), which better enables visualization of these multicellular biofilm layers. While 10 µm spatial resolution is achievable, it is often at the expense of sensitivity. This limitation has been addressed via various enrichment technologies, such as continuous accumulation of selected ions (CASI) [49] or MALDI-2 postionization [50], [51]. However, the utility of each of these enrichment technologies can present new challenges, such as limited mass range and spatial resolution for ion accumulation technologies, or increased ion suppression by chemical interferences produced in the post-ionization process. Incorporation of ion mobility separation prior to mass analysis has also been demonstrated to increase sensitivity by reducing chemical noise and increasing peak capacity [52] and S/N [53], [54] in an IMS experiment. Ion mobility utilizes opposing forces of an inert gas to direct analytes in one direction and a voltage gradient directing analytes in the opposite direction to achieve molecular separation as a function of the size-to-charge ratio of an analyte [57], [58]. Numerous types of ion mobility have been integrated with IMS including drift-tube [59], [60], travelling-wave [53], high-field asymmetric waveform [61], and we recently reported the integration of trapped ion mobility spectrometry (TIMS) with IMS [52], [62]. Through the implementation of TIMS for increased sensitivity, here we perform 10 µm spatial resolution MALDI IMS of cross-sections for the investigation of lipids within distinct, horizontal S. aureus biofilm layers to uncover stark inter-layer heterogeneity at the lipidomic level.
Methods
Materials
2′,5′-Dihydroxyacetophenone (DHA), 1′,5′-Diaminonaphthalene (DAN), ammonium formate, ammonium acetate, and poly-l-lysine solution were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). HPLC-grade acetonitrile, methanol, and gelatin powder were purchased from Fisher Scientific (Pittsburgh, PA, USA). Carboxymehtylcellulose (CMC) was purchased from PM MP Biomedicals (Salon, OH). Tryptic soy agar (TSA) and tryptic soy broth (TSB) were purchased from Becton Dickinson (Franklin Lakes, NJ) and all lab plasticware was from USA Scientific (Ocala, FL) or Corning (Corning, NY). Whatman Nucleopore polycarbonate filter discs (0.1 um pore size) were acquired from Millipore Sigma (Burlington, MA).
Sample preparation
S. aureus bacterial cultures and biofilm growth. S. aureus strains USA300 LAC [73] or USA300 LAC modified to constitutively express GFP [74] were streaked on TSA and incubated at 37 °C. Single colonies were picked and used to inoculate 5 mL of TSB in 15 mL polypropylene tubes. Tubes were agitated for 16 hr at 37 °C. Twenty microliters of the resulting stationary phase culture were then pipetted onto filter discs on TSA plates and incubated at 37 °C for a total of 72 h. During this time, filter discs were transferred to fresh TSA plates every 24 h. Biofilm transfers were performed using antiseptic technique with disinfected forceps to grasp the edge of the filter disc, with care to maintain the biofilm in a horizontal orientation, and to not make contact with the biofilm. In order to measure S. aureus biofilm growth over time CFU counts were obtained every 24 h after inoculation. To measure CFUs at the appropriate time points individual filter discs containing biofilms were placed into 1.5 mL Navy Eppendorf Bead Lysis Kit tubes containing 1 mL of Phosphate Buffered Saline and homogenized at 4° C for three successive 4 min cycles using a Bullet Blender Tissue Homogenizer (Next Advance, Inc., Troy, NY) on the maximum speed. Serial dilutions of the homogenates were plated on TSA to determine CFUs.
Anaerobic biofilms were grown by inoculating filters discs as described above and then placing the petri dishes containing the filter discs in a 37 °C incubator housed inside an anaerobic chamber (Coy Lab Products, Inc., Grass Lake, MI) filled with a mixture of 90 % nitrogen, 5 % hydrogen, 5 % carbon dioxide. Filter discs were transferred every 24 h to fresh TSA plates that had been allowed to equilibrate to the anaerobic chamber for at least 24 h prior to transfer.
After 72 h growth, S. aureus biofilms were embedded in a CMC/gelatin mix and frozen at −80 °C. Samples were then cryosectioned to a 10 μm thickness using a CM3050 S cryostat (Leica Biosystems, Wetzlar, Germany), and biofilm sections were then transferred to a conductive indium tin oxide (ITO) coated glass slide (Delta Technologies, Loveland, CO) coated with poly-l-lysine. A Teflon-coated slide was used to press down on top of the biofilm section, sandwiching it between the ITO slide and the Teflon-coated slide, resulting in even adherence of the section to the ITO slide. Sections were then thaw-mounted as normal. Washing was performed using an eyedropper 3X for 30 s and dried in a vacuum desiccator. Autofluorescence and fluorescence microscopy images were acquired using enhanced green fluorescent protein (EGFP), filter when imaging only green fluorescent protein (GFP), or EGFP, 4′,6-diamidino-2-phenylindole (DAPI), and red fluorescent protein from Discosoma (DsRed) filters for autofluorescence on a Zeiss AxioScan.Z1 slide scanner (Carl Zeiss Microscopy GmbH, Oberkochen, Germany) before matrix application. Briefly, 150 mg of 2′,5′-Dihydroxyacetophenone (DHA) was dissolved in 10 mL of a methanol/acetonitrile/water (80:10:10) solution and applied to a density of 2.08 µg/mm2 by an M5 TM sprayer (HTX Technologies, Chapel Hill, NC). 1′,5′-Diaminonaphthalene (DAN) was applied similarly by dissolving 100 mg into acetonitrile/water (90:10) and spraying to a density of 1.67 µg/mm2.
Imaging mass spectrometry
All experiments were performed on a prototype timsTOF fleX mass spectrometer (Bruker Daltonik, Bremen, Germany) [52]. Images were acquired in TIMS mode of operation with an ion transfer time of 110 μs and a prepulse storage time of 5 μs. Biofilm imaging data were collected using the Bruker Smartbeam 3D laser capable of steering the laser beam to ablate an adjustable square area, termed beam scanning. The beam scan was set to 6 × 6 µm with a pitch of 10 μm using 70 shots per pixel and 59 % laser power. MALDI IMS burn patterns are provided to demonstrate the resultant laser ablation (Supplemental Figure 14). Positive and negative ionization mode data were collected on biofilm serial sections scanning from m/z 300 – 1,300 and m/z 100 – 2000, respectively. The TIMS EFG scan time was set to 500 ms, with a tunnel-in pressure of 2.1 mbar and reduced mobility (1/K0) range of 0.8–2.2 (V·s)/cm2. TIMS imaging data were visualized via SCiLS Lab (Bruker Daltonik, Bremen, Germany) where hotspot removal was applied. In brief, each signal intensity scale was normalized to the maximum intensity detected for a given ion and the color map is extended for enhanced perception of texture and dynamic range. k-means clustering was also performed in SCiLS using total ion current (TIC) normalization, k = 5 and a correlation distance metric. Lipid identifications were determined with high mass accuracy and LIPIDMAPS lipidomics gateway (lipidmaps.org) [75], [76] and confirmed by MS/MS fragmentation where possible. Experiments for lipid abundance comparisons were run in technical triplicate and violin plots were generated using in-house developed software capable of considering both m/z and TIMS dimensions of the data.
Data processing
The data were exported into a custom binary format optimized for storage and speed of analysis of the ion mobility-IMS data. Each frame/pixel contains 104-106 centroid peaks that span the acquisition range of m/z 100 – 2000 with 443,878 and 3,818 bins in the MS and ion mobility dimensions, respectively. The individual centroid peaks were inserted at their correct bin position along the MS and ion mobility dimensions and missing values were set to zero. Data was mass aligned using seven automatically selected m/z features and mass calibrated to several known standard molecules. The datasets were also ion mobility aligned to remove any drift time shift acquired during the experiment. Ion mobility-aware ion images were extracted using a ± 3 ppm mass window around each studied mass to produce a matrix of the number of pixels * the number of ion mobility bins. Such images can be used to create an ion image (summing all mobility information into a single vector), a subset of mobility bins can be selected (e.g., to select one or multiple conformations) or all pixels can be summed to produce an extracted ion mobilogram (XIM). In all cases, ion mobility images are normalized using TIC normalization.
Statistics
The XIM was fitted with a Gaussian curve to determine the full-width half maximum (FWHM) of the single conformation of the ion. The FWHM boundaries were used to select a subset of ion mobility bins within the ion mobility image and the 2D matrix was summed along the ion mobility dimension to produce a vector of intensities. The top and bottom masks were used to sub-select a set of pixels in the vector of intensities; each subset of pixels was then collected into a table and padded with NaNs to make sure the two vectors were identical in size and shape. Padding was necessary due to slight differences in the number of pixels in the top and bottom layer masks. All statistics were computed while excluding NaNs using the pandas Python package while the violin plots were generated using the seaborn visualization library.
Results and discussion
Biofilm growth and sample preparation for IMS
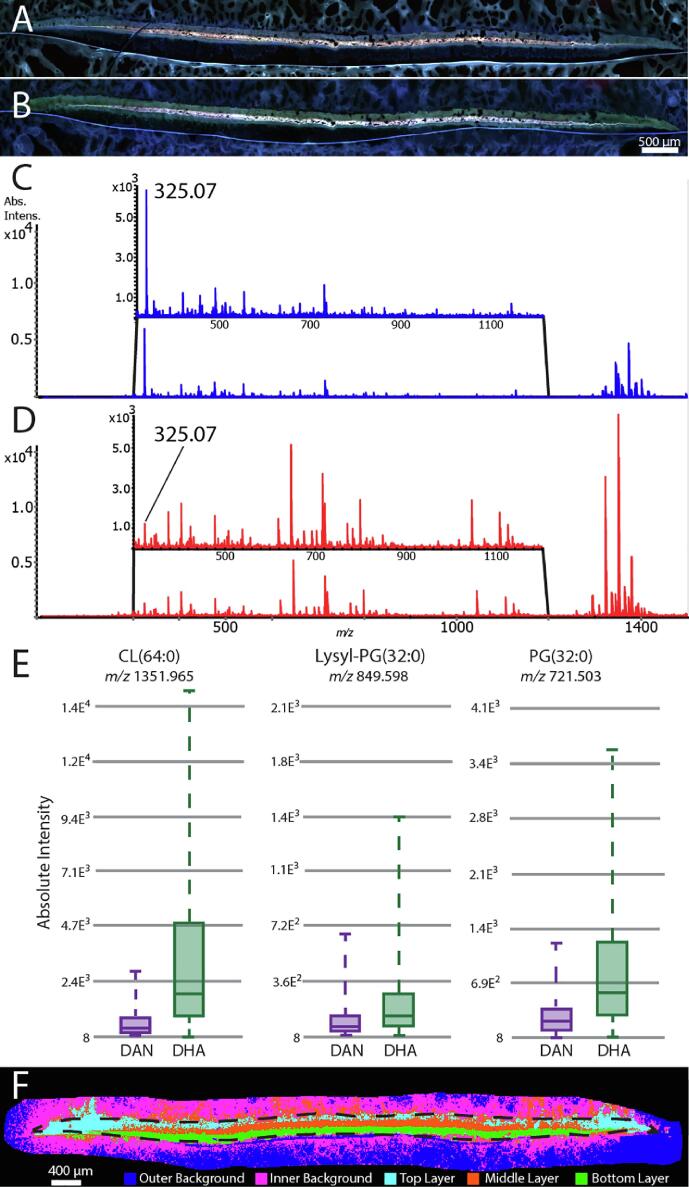
S. aureus biofilm preparation consisted of growth, embedding, cryosectioning, washing, autofluorescence microscopy, matrix deposition, and MALDI IMS analysis (Fig. 1). S. aureus biofilms were grown on tryptic soy agar (TSA) for 72 hrs to generate biofilms of ∼ 400 µm thickness. Biofilms grown in this manner reproducibly reached maturity by 24 h in terms of the number of bacteria, as counted by colony forming units (CFUs), and maintained a constant CFU count over the next 48 h (Supplemental Fig. 1A). Optical images of biofilms were acquired at each transfer timepoint to provide additional morphological context throughout development (Supplemental Fig. 1B). Unlike other bacterial species such as P. aeruginosa [63], S. aureus biofilms grown in vitro often lack structural rigidity, making it challenging to retain their morphology during IMS sample preparation and necessitating embedding of the biofilm. Traditional embedding materials, such as OCT, interfere with MALDI IMS analysis. As a result, we elected to embed the biofilms in a mixture of CMC and gelatin (5 % and 10 % w/w, respectively) to provide structural support during cryosectioning [64]. Notably, we found that adhering the biofilm section evenly to the microscope slide by pressing down with a Teflon-coated microscope slide before thaw-mounting was critical for maintaining biofilm morphology with discrete layers (Supplemental Fig. 2A). Washing with ammonium formate and ammonium acetate solutions has been previously reported for MALDI IMS of tissue [65], but has not been investigated for use in S. aureus biofilms to our knowledge. For these reasons, we assessed HPLC grade water and both 50 mM and 150 mM solutions of ammonium formate and ammonium acetate for removing the salt and other additives present in growth media (Supplemental Fig. 2B). Sample loss resulting from each wash procedure was determined by autofluorescence microscopy by comparing before and after wash images. Of all the washes, 150 mM ammonium formate minimally perturbed sample morphology (Fig. 2A and 2B), while water caused the most damage. Furthermore, we found that washing with 150 mM ammonium formate yielded increased lipid signal for lipid species above m/z 300, especially enhancing CL lipid signal (Fig. 2C and 2D) compared to unwashed samples. For example, CL(64:0) displayed a more than tenfold enhancement after washing when analyzed in quadruplicate (Supplemental Fig. 3A – B). The identity of the predominant peak measured at m/z 325.07 in the unwashed samples is unknown, however it localizes to the matrix around the biofilm and primarily to the filter disc. It is likely a chemical interference from the bacterial culture or sample preparation process being remediated by the washing step (Supplemental Fig. 2C). For lipid analysis, we assessed both DAN and DHA matrices as they enable dual polarity imaging, thereby increasing the number and type of lipid species that can be detected. For effective comparison, PG(32:0), L-PG(32:0) and CL(64:0) lipids were selected as representative of three unique and biologically important lipid classes. Absolute signal intensity for each of these lipids was compared for both MALDI matrices and in each case, DHA provided higher signal intensity than DAN (Fig. 2E). Moreover, DAN matrix clusters were detected across the measured m/z range in both polarities (Supplemental Fig. 2D), increasing overall background and conflating with lipid species. For example, lysyl-PG(30:0) was detected with an interfering and overlapping ion using DAN, but fully resolved using DHA (Supplemental Fig. 4A - B). To assess the effect of this on downstream lipid identifications, a LipidMAPS search of these datasets was performed comparing DAN and DHA where the number of annotated peaks using DHA was 388 compared to only 185 when DAN was used. For these reasons, DHA was selected for all future experiments.
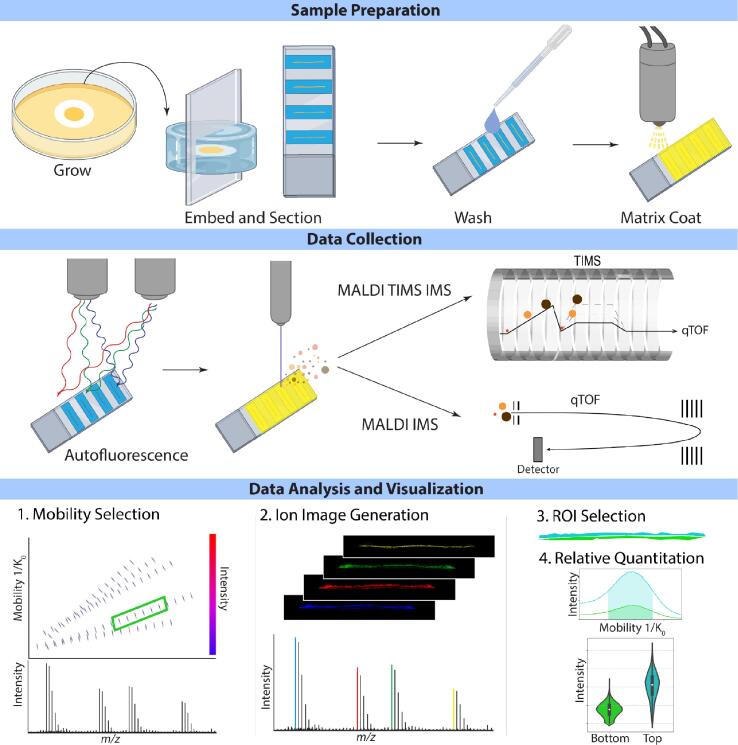
Fig. 1.
Workflow of methodology for IMS of S. aureus biofilms including sample preparation demonstrating the orientation of the biofilm cross-section followed by data collection of both autofluorescence and IMS and finally data analysis and visualization where data are first filtered by TIMS mobility selection. Ion images are created, and relative quantitation is performed between layered subpopulations within the biofilm.
Fig. 2.
Washing of S. aureus biofilms and MALDI matrix selection enhance lipid signal abundance in an IMS experiment. Autofluorescence microscopy shows minimal sample loss before (A), and after (B) washing with 150 mM ammonium formate. Negative mode mass spectra before (C), and after (D) washing show that washing also enhances signal of lipids, while reducing signal of chemical noise. Signal intensity box plots of CL(64:0), L-PG(31:0) and PG(32:0) (E) compare DHA and DAN matrices, demonstrating higher signal intensity for each lipid with DHA. k-means clustering performed on IMS. data of a biofilm demonstrates biofilm layers cluster independently (F).
With the goal of visualizing lipidomically distinct layers within the biofilms, we next collected 10 µm spatial resolution IMS data of an S. aureus biofilm and performed segmentation by k-means clustering (Fig. 2F). This analysis provided an unsupervised approach for clustering pixels whose spectral profiles are similar and then segmenting the imaging data by color coding each pixel by its cluster membership. Using five clusters (k = 5), we found that the matrix background clustered into two classes (outer and inner background, outside dotted line, Fig. 2F). We attributed this independent clustering of outer and inner background to slight delocalization of bacterial molecules or whole bacteria around the main structure of the biofilm. Interestingly, we observed three distinct layers within the biofilm to cluster independently, suggesting that the lipidomic profiles differ significantly in these distinct regions.
We elected to improve sensitivity and coverage of detected lipids using ion mobility. The contributions of TIMS were assessed by comparing peak capacity and signal-to-noise (S/N) with, and without TIMS separation implemented. To do this, we averaged the spectrum of a 10 µm spatial resolution imaging experiment without considering the TIMS dimension, resulting in the detection of 550 features (Supplemental Fig. 5C). Low signal intensity for various lipids, such as L-PG species was observed with S/N as low as 3. Considering the TIMS dimension however, S/N was increased for most detected lipid species. For example, lysyl-PG(28:0) (m/z 793.535) was detected with a S/N of 88 (Supplemental Fig. 5A), but by excluding noise at 1/K0 values, other than the lipid of interest, we found that the S/N improved by more than twofold (S/N 193, Supplemental Fig. 5B). Furthermore, when the TIMS dimension was considered for the dataset as a whole, ion mobility separation revealed ∼ 1,000 additional detected features (1,545), likely consisting of isobars, isomers, and conformers of detected analytes (Supplemental Fig. 5D). Chemical specificity was also enhanced by TIMS, as we found that many ions of interest were observed with notable peak tailing in the TIMS dimension, which could be due to slight separation of additional confounding species from the ion of interest at similar m/z. By selecting a narrow window (0.015) around the peak of interest (Supplemental Fig.6), extraneous species were excluded to enhance chemical specificity. As a result, all downstream analyses were performed with TIMS to reduce the overall spectral noise and increase chemical specificity, and all ion images depict ions within a 1/K0 and m/z range.
S. aureus biofilm imaging
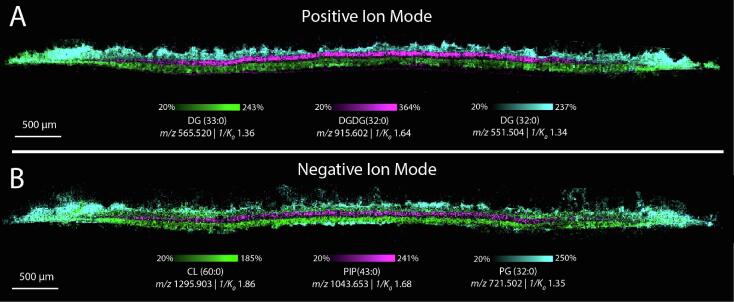
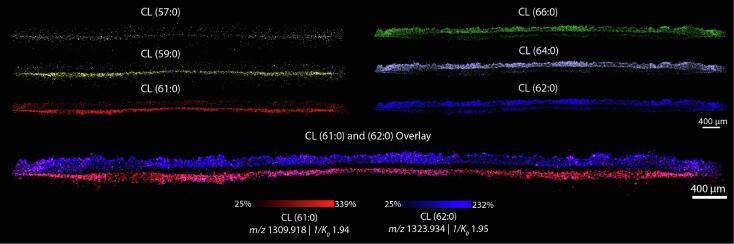
Biofilms are known to comprise distinctive layers with inter-layer heterogeneity in gene regulation [8]. Here, we aimed to assess the lipid distributions within these layers. This growth pattern is thought to arise from a series of steps including attachment, multiplication, and detachment that S. aureus undergoes during the biofilm generation process [8], [66]. Due to their small size, high spatial resolution IMS is required to obtain the necessary spatial fidelity for investigating these discrete layers. We performed 10 µm spatial resolution IMS in both positive and negative ion modes. TIMS was utilized to maximize sensitivity and molecular coverage. Overall, we detected both odd-chain and even-chain PG, Lysyl-PG and CL, as well as phosphatidylcholine (PC), phosphatidylinositol-phosphate (PIP), diacylglycerols (DG) and digalactosyldiacylglycerols (DGDG), underscoring the sensitivity of our method. Furthermore, the spatial and molecular specificity of IMS uncovered three discrete layers within the biofilm. While horizontal S. aureus biofilm layers are known to exist, these results are among the first to visualize this gross structural information on the lipidomic level. As an example, DG(33:0) (m/z 565.520, 1/K0 1.36), DG(32:0) (m/z 551.504, 1/K0 1.34), and DGDG(32:0) (m/z 915.602, 1/K0 1.64) localize to the bottom, top and middle layers, respectively (Fig. 3). Notably, DG(32:0) and DG(33:0) localize to opposite layers of the biofilm, despite differing only by the addition of a single methyl group on the acyl chain, signifying that lipid localization is not based solely on lipid class. Similar trends were observed in other classes of bacterial lipids such as cardiolipins, as discussed below. We also found discrete localizations of lipids to specific biofilm layers in negative ion mode. For example, CL(60:0) (m/z 1295.903, 1/K0 1.86), PIP(43:0) (m/z 1043.653, 1/K0 1.68), and PG(32:0) (m/z 721.502, 1/K0 1.35) localized to the bottom, middle and top layers, respectively. All ions depicted were putatively identified by accurate mass measurement and MS/MS fragmentation where possible (Table S1 and Supplemental Figure 7). More lipid classes ionize in negative ion mode, contributing to the higher diversity of lipids detected. Comparable to previous studies performed via liquid-chromatography tandem mass spectrometry (LC-MSMS) [16], at least five lipid classes were detected in negative ion mode with most ions localizing primarily to a single layer. The lipidomic profile within each of these regions is distinct, and the delineation between each region is visually clear. These results coincide with other work on B. subtilis that demonstrated bacteria in different phenotypic states in varying locations within a biofilm [39], [67], and suggest that S. aureus biofilm layers also act as discrete communities with a complex system of lipid distribution between them. We again found spatial heterogeneity of lipids from within the same class, such as CL(57:0), CL(59:0) and CL(61:0) (m/z 1253.853, 1/K0 1.88, m/z 1281.887, 1/K0 1.91, and m/z 1309.918, 1/K0 1.94), that are all observed primarily in the bottom layer of the biofilm (Fig. 4). Conversely, CL(62:0), CL(64:0) and CL(66:0) (m/z 1323.934, 1/K0 1.95, m/z 1351.965, 1/K0 1.98 and m/z 1379.996, 1/K0 2.01) were all observed primarily in the top layer of the biofilm. All CL lipids were confidently identified by MS/MS (Supplemental Figure 7B – G) (except CL(57:0) due to low signal intensity). If these spatial differences were only observed between different classes of lipid such as PG and CL lipids, this phenomenon might be the result of different growth phases between top and bottom layers. PG lipids are the predominant species in growing S. aureus, switching to CL lipids in the stationary phase [68], [69]. However, there are no known functional differences for species within the cardiolipin class, potentially suggesting distinct functions of CL species within a spatial context. Interestingly, CL signal was below the limit of detection in the middle layer of the biofilm. To determine if this is due to a difference in the cell density in that layer, a biofilm was grown with a strain of USA300 LAC that was genetically modified to constitutively express GFP. We then collected fluorescence microscopy and MALDI TIMS IMS on the same cross-section and found lower GFP signal in the middle layer by fluorescence microscopy, and corresponding absence of CL signal in the same region by MALDI TIMS IMS (Supplemental Figure 8). These results suggest that the absence of CL signal observed in the middle layer may be due to fewer bacteria present in that region of the biofilm. Most odd-chain CL lipids localize primarily to the bottom layer of the biofilm, and even-chain CL lipids are more abundant in the top layer (Fig. 4). However, the trend did not hold in every case in that CL(58:0) and CL(60:0) were observed primarily in the bottom layer of the biofilm (Supplemental Figure 9A and Fig. 3B). Cardiolipin chain length does not fully explain the separation observed between biofilm layers. Although it appears that longer chain length cardiolipins localize to the top layer and shorter chain length to the bottom, several cardiolipins with longer acyl chain length than 61:0, such as 65:0 have similar abundances between top and bottom layers (Supplemental Figure 9B). While the biological drivers of these observations are still unclear, these results show that CL lipids differing by only one methyl group have starkly different localizations within the microbial colony and further underscores the complexity of the lipid distribution patterns between layers within S. aureus biofilms.
Fig. 3.
High spatial resolution IMS (10 µm) collected from a cross-section of an S. aureus biofilm in positive ion mode (A) and negative ion mode (B) showing delineation of three distinct layers. DG(33:0) (m/z 565.520,1/K0 1.36, 1.77 ppm error) (and CL(60:0) (m/z 1295.903, 1/K0 1.86, 0.46 ppm error) appear to localize primarily to the bottom layer, while DGDG(32:0) (m/z 915.64, 1/K0 1.637, 0.55 ppm error) and PIP(43:0) (m/z 1043.653, 1/K0 1.68, 3.83 ppm error) localize to the middle layer. Finally, DG(32:0) (m/z 551.504, 1/K0 1.34, 1.09 ppm error) and PG(32:0) (m/z 721.502, 1/K0 1.35, 0.14 ppm error) are more abundant in the top layer.
Fig. 4.
Bacterial cardiolipin species distribute heterogeneously within biofilm layers. CL(57:0), CL(59:0) and CL(61:0) distribute primarily to the bottom layer, despite being from the same lipid class. Similarly, CL(62:0), CL(64:0) and CL(66:0) distribute primarily to the top layer of the biofilm. An overlay of CL(61:0) and CL(62:0) clearly demonstrates the delineation.
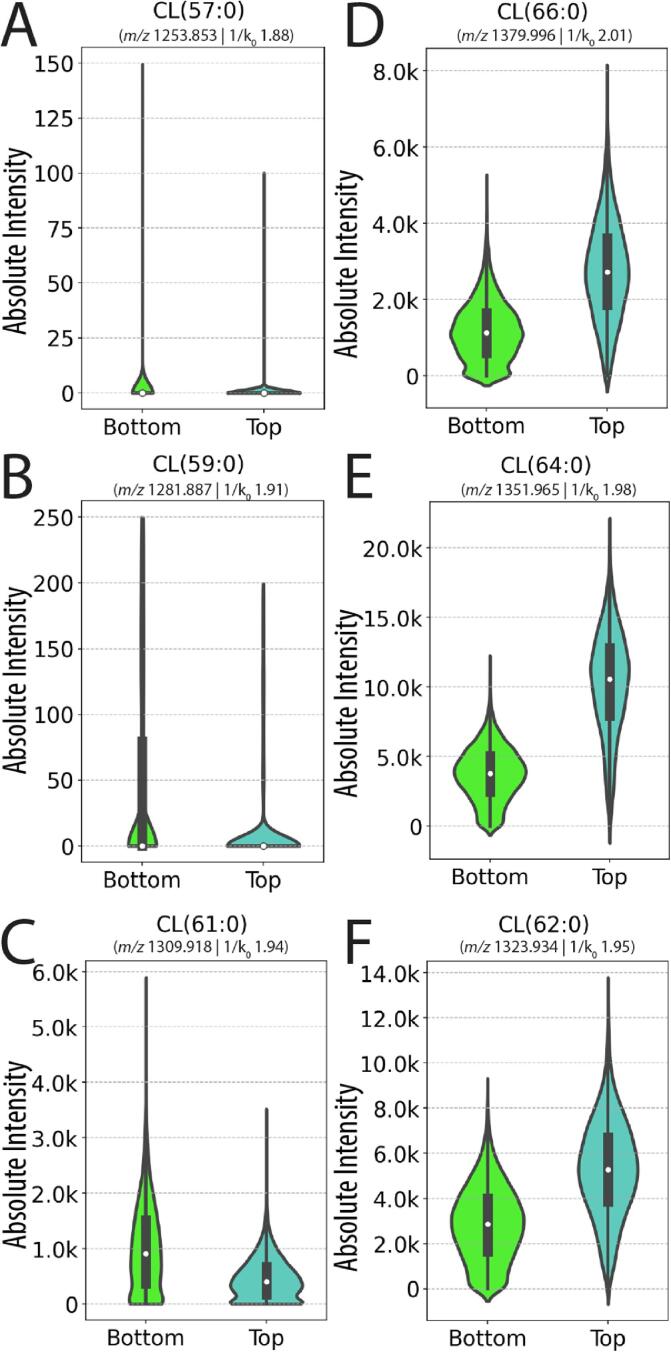
We wanted to further assess the heterogeneity of lipidomic profiles between biofilm layers by making relative comparisons of lipid abundances between them. By acquiring IMS of a biofilm in triplicate, we first extracted spectra from regions of interest of top and bottom layers (Supplemental Fig. 6A–C). After selecting ion mobility (1/K0) windows (Supplemental Fig. 6A–C) to remove chemical noise and any extraneous species, we compared mean signal intensities for each CL species shown in the ion images above (Fig. 5 and Supplemental Figure 10A – B). We found that CL(57:0) CL(59:0) and CL(61:0) were 3.7-, 6.3-, and 2.3-fold more intense in the bottom layer than in the top layer (Fig. 5A - C and Supplemental Figure 10A - B). In contrast, CL(62:0), CL(64:0), and CL(66:0), which primarily localize to the top layer, demonstrated a 1.8, 2.6, and 2.2-fold difference between layers (Fig. 5D - F and Supplemental Figure 10A - B). These findings are also clear in the extracted spectra from each layer (Supplemental Figure 11A – F). The means for these six ions were significantly different (n = 3, p < 0.03) between the top and the bottom layers. In addition to the quantitative analysis performed in technical triplicate, we confirmed that these trends were consistent in biological replicate (USA300 LAC), as well as a different isolate of USA300 (JE2), (Supplemental Figure 12A – B). Although this analysis is an example that analyzes only one class of bacterial lipid species, it demonstrates that the lipidomic differences between biofilm layers are significant and further supports that they are distinct at the lipidomic level.
Fig. 5.
S. aureus biofilms exhibit lipidomic heterogeneity between layers. Violin plots generated from one of three replicate images show that CL(57:0), CL(59:0) and CL(61:0) are more abundant in the spectrum extracted from the bottom layer (green) when compared to the top layer (cyan) (A - C), and that CL(62:0), CL(64:0) and CL(66:0) are more abundant in the top layer compared to the bottom (D – F). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
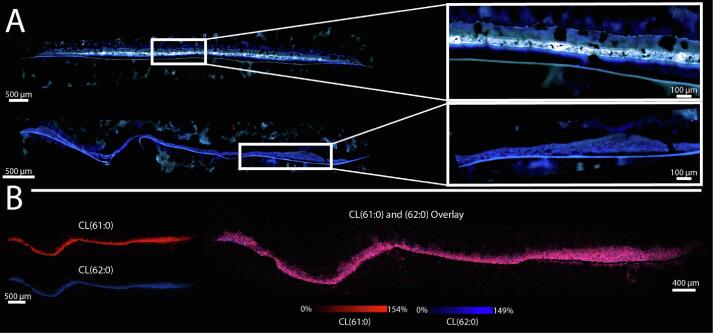
This pronounced lipidomic heterogeneity between S. aureus biofilm layers led us to probe the biological drivers of this phenomenon. Notably, S. aureus is a facultative anaerobe with the ability to survive in both aerobic and anaerobic conditions [70] S. aureus growth under anaerobic conditions leads to significant differences in fatty acid content. Specifically, anaerobic metabolism results in a shift from long to short isobranched fatty acids [71]. Changes in fatty acids, a major building block of lipids, could result in a shift in lipid content, as each observed lipid class is synthesized from the same pool of available fatty acids. We hypothesized that these layered subpopulations experience different amounts of accessible oxygen, and resultant shifts in fatty acid content could explain differences in CL lipid content between them. We investigated the role of oxygen in the spatial distributions of lipids in biofilms grown under anaerobic conditions compared to those grown under atmospheric conditions for the same duration. To investigate the growth of aerobic vs anaerobic S. aureus biofilms, we compared CFUs between anaerobic biofilms to their aerobic counterparts during biofilm development. At 72 hrs, when the MALDI TIMS IMS was performed, biofilms grown in both conditions had reached densities greater than 1x109 CFUs per filter. While significantly different from one another, the difference between CFUs in the two conditions was still less than half a log (Supplemental Fig. 1A). Furthermore, after 24 hrs of development, both the aerobic and anaerobic biofilm populations were quite stable with no significant changes in their number of CFUs between 48 and 72 hrs. Autofluorescence of an anaerobic biofilm revealed considerably thinner single-layered biofilms, rather than three distinct layers observed in aerobic biofilms (Fig. 6A). We collected IMS data and performed k-means clustering with no discernable layers clustered separately in the IMS data. Instead, the entire biofilm clustered together, further suggesting the existence of a single layer (Supplemental Figure 13). The absence of layers indicates that oxygen may be necessary to induce S. aureus biofilm layer generation that could have implications for biofilm growth in environments of varying oxygen availability. In particular, if the layers observed in aerobic biofilms are the result of waves of bacterial detachment, a known step of the S. aureus biofilm generation process [66], [72], these results suggest that oxygen may be required for biofilm detachment. Our findings also align with the hypothesis that there may exist a mechanism of biofilm structure formation involving metabolic differentiation of cells leading to metabolically distinct subpopulations within an S. aureus biofilm [66]. If this process depends on the presence of oxygen, this could lead to a lack of bacterial differentiation and subsequent absence of biofilm layers. In addition to the loss of layered subpopulations, we observed homogeneous lipid distributions spanning the cross-section of the anaerobic biofilm. For example, when comparing ion images of CL(61:0) and CL(62:0), both species appear to share the same spatial distribution across the entire biofilm (Fig. 6B). This is in contrast to that of the aerobic biofilm where these lipid species demonstrated distinct localizations to discrete biofilm layers (Fig. 4). We observed all CL species in the anaerobic biofilm, indicating that oxygen is not necessary for the production of these lipid species. However, this does not rule out that an oxygen gradient could be contributing to heterogeneity of lipid abundances between biofilm layers in aerobic biofilms.
Fig. 6.
S. aureus biofilms grown under anaerobic conditions generate thinner biofilms (bottom) comprised of a single layer compared to those grown under aerobic conditions (top) (A). IMS of a biofilm grown under anaerobic conditions reveals homogenous distribution of CL(61:0) and CL(62:0) (B) which have different distributions in biofilms grown under aerobic conditions.
Conclusions
Various sample preparation protocols including embedding in a mixture of CMC/gelatin and isotonic washing with ammonium formate resulted in preservation of S. aureus biofilm structure while removing spectral interferences. IMS and k-means clustering yielded unsupervised visualization of S. aureus biofilm layers, providing an initial indication of the lipidomic heterogeneity between biofilm layers. Gas-phase separation by TIMS also contributed to increasing both the S/N and the overall peak capacity in 10 µm spatial resolution IMS. Discrete, lipidomically distinct layered subpopulations were revealed by IMS of bacterial lipids in both positive and negative polarities facilitated by the methodologies developed herein. Not only were different lipids observed to localize primarily to discrete layers, but lipids from within the same class, in many cases only differing by one methyl group on the acyl chain, were observed to localize primarily to different layers. Relative quantitation from extracted mass spectra of discrete layers afforded inter-layer comparison of individual lipid abundances and demonstrated differences in select CL lipid species between layers. Further, our findings suggest that individual CL’s that have historically been thought to contribute cooperatively toward certain functions, could be playing distinct biological roles. Future work growing S. aureus biofilms with access to defined fatty acids, or genetic alterations of various enzymes involved in CL synthesis could elucidate the biological drivers of these lipid distributions. Following the CL study, we found that S. aureus biofilms grown under anaerobic conditions developed thinner biofilms comprised of a single layer compared to thicker biofilms comprised of three layers when grown under aerobic conditions. This suggests that the generation of multiple layers in S. aureus biofilms may require aerobic growth conditions. Given that biofilms grown anaerobically are not able to reach the same CFU’s as those grown under aerobic conditions, it remains possible that these differences are due to differences in size of the biofilm as opposed to oxygen concentration. However, because both growth conditions reached CFU’s that remained stable over time, we would not expect development of horizontal layers in anaerobic biofilms over an extended time period. IMS of these anaerobic biofilms revealed that the lipids analyzed in this work localized homogeneously throughout the anaerobic biofilm. Although further analyses are required to fully understand the significance of lipid distributions within S. aureus biofilms, this work using IMS technology demonstrates significant lipidomic heterogeneity between biofilm layers. Furthermore, the methodologies developed herein have the potential to be applied towards understanding spatially discrete biofilm responses to external stimuli, such as exposure to antimicrobials. Studies of this nature could reveal localized molecular characteristics that could be exploited for new therapeutic strategies.
Associated content
The raw data are available at https://doi.org/10.6084/m9.figshare.c.6054968.v1.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Support was provided by The National Institutes of Health (NIH) National Institute of Allergies and Infectious diseases (NIAID) (R01AI138581 awarded to J.M.S. and E.P.S), and The National Science Foundation Major Research Instrument Program (CBET—1828299 awarded to J.M.S. and R.M.C.). J.A.F. was supported by a T32 training grant from the NIH NIAID (T32 AI007474). E.K.N. was supported by a National Institute of Environmental Health Sciences training grant (T32ES007028). Fig. 1 was created with BioRender.com (Agreement number EW22X0L5U9).
Footnotes
Peer review under responsibility of “MSACL”.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmsacl.2022.09.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Giraudy C., Hausmann S., Lemeille S., Prados J., Redder P., Linder P. The C-terminal region of the RNA helicase CshA Is required for the interaction with the degradosome and turnover of bulk RNA in the opportunistic pathogen staphylococcus aureus. RNA Biol. 2015;12(6):658–674. doi: 10.1080/15476286.2015.1035505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kourtis A.P., Hatfield K., Baggs J., Mu Y., See I., Epson E., Nadle J., Kainer M.A., Dumyati G., Petit S., Ray S.M., Ham D., Capers C., Ewing H., Coffin N., McDonald L.C., Jernigan J., Cardo D. Vital Signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus Aureus bloodstream infections — United States. MMWR Morb Mortal Wkly Rep. 2019;68(9):214–219. doi: 10.15585/mmwr.mm6809e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otto, M. Staphylococcal Biofilms. Current Topics in Microbiology and Immunology. Curr Top Microbiol Immunol 2008, pp 207–228. https://doi.org/10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed]
- 4.Rohde H., Burandt E.C., Siemssen N., Frommelt L., Burdelski C., Wurster S., Scherpe S., Davies A.P., Harris L.G., Horstkotte M.A., Knobloch J.K.M., Ragunath C., Kaplan J.B., Mack D. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of staphylococcus epidermidis and staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28(9):1711–1720. doi: 10.1016/J.BIOMATERIALS.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 5.Balaban N., Cirioni O., Giacometti A., Ghiselli R., Braunstein J.B., Silvestri C., Mocchegiani F., Saba V., Scalise G. Treatment of Staphylococcus aureus biofilm infection by the quorum-sensing inhibitor RIP. Antimicrob Agents Chemother. 2007;51(6):2226–2229. doi: 10.1128/AAC.01097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Römling U., Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;272(6):541–561. doi: 10.1111/JOIM.12004. [DOI] [PubMed] [Google Scholar]
- 7.Bernier S.P., Lebeaux D., DeFrancesco A.S., Valomon A., Soubigou G., Coppée J.-Y., Ghigo J.-M., Beloin C., Viollier P.H. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 2013;9(1):e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Periasamy S., Joo H.S., Duong A.C., Bach T.H.L., Tan V.Y., Chatterjee S.S., Cheung G.Y.C., Otto M. How staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A. 2012;109(4):1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Resch A., Rosenstein R., Nerz C., Götz F. Differential gene expression profiling of staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol. 2005;71(5):2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Lopez M.I., Mendez-Reina R., Trier S., Herrfurth C., Feussner I., Bernal A., Forero-Shelton M., Leidy C. Variations in carotenoid content and acyl chain composition in exponential, stationary and biofilm states of staphylococcus aureus, and their influence on membrane biophysical Properties. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2019;1861(5):978–987. doi: 10.1016/J.BBAMEM.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Slavetinsky C.J., Peschel A., Ernst C.M. Alanyl-phosphatidylglycerol and lysyl-phosphatidylglycerol are translocated by the same mprf flippases and have similar capacities to protect against the antibiotic daptomycin in staphylococcus aureus. Antimicrob Agents Chemother. 2012;56(7):3492–3497. doi: 10.1128/AAC.00370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilelee E., Pokorny A., Yeaman M.R., Bayer A.S. Lysyl-phosphatidylglycerol attenuates membrane perturbation rather than surface association of the cationic antimicrobial peptide 6W-RP-1 in a model membrane system: implications for daptomycin resistance. Antimicrob Agents Chemother. 2010;54(10):4476–4479. doi: 10.1128/AAC.00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J.-H., Bhuiyan M.S., Shen H.-H., Cameron D.R., Rupasinghe T.W.T., Wu C.-M., Le Brun A.P., Kostoulias X., Domene C., Fulcher A.J., McConville M.J., Howden B.P., Lieschke G.J., Peleg A.Y. Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc. Natl. Acad. Sci. U.S.A. 2019;116(9):3722–3727. doi: 10.1073/pnas.1812066116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn S., Slavetinsky C.J., Peschel A. Synthesis and function of phospholipids in staphylococcus aureus. Int. J. Med. Microbiol.. Elsevier GmbH February 1. 2015;305(2):196–202. doi: 10.1016/j.ijmm.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Tsai M., Ohniwa R.L., Kato Y., Takeshita S.L., Ohta T., Saito S., Hayashi H., Morikawa K. Staphylococcus aureus requires cardiolipin for survival under conditions of high salinity. BMC Microbiol. 2011;11(1) doi: 10.1186/1471-2180-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hines K.M., Waalkes A., Penewit K., Holmes E.A., Salipante S.J., Werth B.J., Xu L., Limbago B.M. Characterization of the mechanisms of daptomycin resistance among gram-positive bacterial pathogens by multidimensional lipidomics. mSphere. 2017;2(6) doi: 10.1128/msphere.00492-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohlenkamp C., Geiger O., Narberhaus F. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol Rev. 2016;40(1):133–159. doi: 10.1093/femsre/fuv008. [DOI] [PubMed] [Google Scholar]
- 18.Hines K.M., Alvarado G., Chen X.i., Gatto C., Pokorny A., Alonzo F., Wilkinson B.J., Xu L., Ellermeier C.D. Lipidomic and ultrastructural characterization of the cell envelope of staphylococcus aureus grown in the presence of human serum. Lipidomic and Ultrastructural Characterization of the Cell Envelope of Staphylococcus Aureus Grown in the Presence of Human Serum. 2020;5(3) doi: 10.1128/mSphere.00339-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubio A., Moore J., Varoglu M., Conrad M., Chu M., Shaw W., Silverman J.A. LC-MS/MS Characterization of Phospholipid Content in Daptomycin-Susceptible and -Resistant Isolates of Staphylococcus Aureus with Mutations in MprF. 2012;29(1):1–8. doi: 10.3109/09687688.2011.640948. doi: 10.3109/09687688.2011.640948. [DOI] [PubMed] [Google Scholar]
- 20.O'Brien A.D., Bayer A.S., Prasad R., Chandra J., Koul A., Smriti M., Varma A., Skurray R.A., Firth N., Brown M.H., Koo S.-P., Yeaman M.R. In vitro resistance of staphylococcus aureus to thrombin-induced platelet microbicidal protein is associated with alterations in cytoplasmic membrane fluidity. Infect Immun. 2000;68(6):3548–3553. doi: 10.1128/iai.68.6.3548-3553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M., Rigby K., Lai Y., Nair V., Peschel A., Schittek B., Otto M. Staphylococcus aureus mutant screen reveals interaction of the human antimicrobial peptide dermcidin with membrane phospholipids. Antimicrob Agents Chemother. 2009;53(10):4200–4210. doi: 10.1128/AAC.00428-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornett D.S., Reyzer M.L., Chaurand P., Caprioli R.M. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4(10):828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Yang E., Chaurand P., Raghavan V. Visualizing the distribution of strawberry plant metabolites at different maturity stages by MALDI-TOF imaging mass spectrometry. Food Chem. 2021;345 doi: 10.1016/J.FOODCHEM.2020.128838. [DOI] [PubMed] [Google Scholar]
- 24.Palmer A., Phapale P., Chernyavsky I., Lavigne R., Fay D., Tarasov A., Kovalev V., Fuchser J., Nikolenko S., Pineau C., Becker M., Alexandrov T. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods. 2017;14(1):57–60. doi: 10.1038/nmeth.4072. [DOI] [PubMed] [Google Scholar]
- 25.Powers T.W., Neely B.A., Shao Y., Tang H., Troyer D.A., Mehta A.S., Haab B.B., Drake R.R., Batra S.K. MALDI imaging mass spectrometry profiling of N-Glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS ONE. 2014;9(9):e106255. doi: 10.1371/journal.pone.0106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heijs B., Holst S., Briaire-de Bruijn I.H., van Pelt G.W., de Ru A.H., van Veelen P.A., Drake R.R., Mehta A.S., Mesker W.E., Tollenaar R.A., Bovée J.V.M.G., Wuhrer M., McDonnell L.A. Multimodal mass spectrometry imaging of N-glycans and proteins from the same tissue section. Anal Chem. 2016;88(15):7745–7753. doi: 10.1021/acs.analchem.6b01739. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H., Shi X., Vu N.Q., Li G., Li Z., Shi Y., Li M., Wang B., Welham N.V., Patankar M.S., Weisman P., Li L. On-tissue derivatization with girard’s reagent P enhances N-glycan signals for formalin-fixed paraffin-embedded tissue sections in MALDI mass spectrometry imaging. Anal Chem. 2020;92(19):13361–13368. doi: 10.1021/ACS.ANALCHEM.0C02704/ASSET/IMAGES/LARGE/AC0C02704_0005.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis S.R., Paine M.R.L., Eijkel G.B., Pauling J.K., Husen P., Jervelund M.W., Hermansson M., Ejsing C.S., Heeren R.M.A. Automated, parallel mass spectrometry imaging and structural identification of lipids. Nat Methods. 2018;15(7):515–518. doi: 10.1038/s41592-018-0010-6. [DOI] [PubMed] [Google Scholar]
- 29.Bednařík A., Bölsker S., Soltwisch J., Dreisewerd K. An On-tissue paternò-büchi reaction for localization of carbon-carbon double bonds in phospholipids and glycolipids by matrix-assisted laser-desorption–ionization mass-spectrometry imaging. Angew. Chem. 2018;130(37):12268–12272. doi: 10.1002/ANGE.201806635. [DOI] [PubMed] [Google Scholar]
- 30.Altelaar A.F.M., Taban I.M., McDonnell L.A., Verhaert P.D.E.M., de Lange R.P.J., Adan R.A.H., Mooi W.J., Heeren R.M.A., Piersma S.R. High-resolution MALDI imaging mass spectrometry allows localization of peptide distributions at cellular length scales in pituitary tissue sections. Int J Mass Spectrom. 2007;260(2–3):203–211. doi: 10.1016/J.IJMS.2006.09.028. [DOI] [Google Scholar]
- 31.McDonnell L.A., Corthals G.L., Willems S.M., van Remoortere A., van Zeijl R.J.M., Deelder A.M. Peptide and protein imaging mass spectrometry in cancer research. J Proteomics. 2010;73(10):1921–1944. doi: 10.1016/J.JPROT.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Piga I., Heijs B., Nicolardi S., Giusti L., Marselli L., Marchetti P., Mazzoni M.R., Lucacchini A., McDonnell L.A. Ultra-high resolution MALDI-FTICR-MSI analysis of intact proteins in mouse and human pancreas tissue. Int J Mass Spectrom. 2019;437:10–16. doi: 10.1016/J.IJMS.2017.11.001. [DOI] [Google Scholar]
- 33.Walch A., Rauser S., Deininger S.O., Höfler H. MALDI imaging mass spectrometry for direct tissue analysis: A new frontier for molecular histology. Histochem Cell Biol. 2008;130(3):421–434. doi: 10.1007/S00418-008-0469-9/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caprioli R.M., Farmer T.B., Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez D.J., Haste N.M., Hollands A., Fleming T.C., Hamby M., Pogliano K., Nizet V., Dorrestein P.C. Microbial competition between bacillus subtilis and staphylococcus aureus monitored by imaging mass spectrometry. Microbiology (N Y) 2011;157(9):2485–2492. doi: 10.1099/MIC.0.048736-0/CITE/REFWORKS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez D.J., Okumura C.Y., Hollands A., Kersten R., Akong-Moore K., Pence M.A., Malone C.L., Derieux J., Moore B.S., Horswill A.R., Dixon J.E., Dorrestein P.C., Nizet V. Novel phenol-soluble modulin derivatives in community-associated methicillin-resistant staphylococcus aureus identified through imaging mass spectrometry. J. Biol. Chem. 2012;287(17):13889–13898. doi: 10.1074/JBC.M112.349860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez D.J., Vuong L., Gonzalez I.S., Keller N., McGrosso D., Hwang J.H., Hung J., Zinkernagel A., Dixon J.E., Dorrestein P.C., Nizet V. Phenol soluble modulin (PSM) variants of community-associated methicillin-resistant staphylococcus aureus (MRSA) captured using mass spectrometry-based molecular networking. Mol. Cell. Proteomics. 2014;13(5):1262–1272. doi: 10.1074/MCP.M113.031336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watrous J.D., Phelan V.V., Hsu C.-C., Moree W.J., Duggan B.M., Alexandrov T., Dorrestein P.C. Microbial metabolic exchange in 3D. ISME J. 2013;7(4):770–780. doi: 10.1038/ismej.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yannarell S.M., Veličković D., Anderton C.R., Shank E.A. Direct visualization of chemical cues and cellular phenotypes throughout bacillus subtilis biofilms. mSystems. 2021;6(6) doi: 10.1128/MSYSTEMS.01038-21/SUPPL_FILE/MSYSTEMS.01038-21-SF002.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moree W.J., Phelan V.V., Wu C.H., Bandeira N., Cornett D.S., Duggan B.M., Dorrestein P.C. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc Natl Acad Sci U S A. 2012;109(34):13811–13816. doi: 10.1073/PNAS.1206855109/SUPPL_FILE/SAPP.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frydenlund Michelsen C., Hossein Khademi S.M., Krogh Johansen H., Ingmer H., Dorrestein P.C., Jelsbak L. Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction. ISME J. 2016;10(6):1323–1336. doi: 10.1038/ismej.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Si T., Li B., Zhang K., Xu Y., Zhao H., Sweedler J.V. Characterization of bacillus subtilis colony biofilms via mass spectrometry and fluorescence imaging. J Proteome Res. 2016;15(6):1955–1962. doi: 10.1021/ACS.JPROTEOME.6B00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phelan V.V., Fang J., Dorrestein P.C. Mass spectrometry analysis of pseudomonas aeruginosa treated with azithromycin. J Am Soc Mass Spectrom. 2015;26(6):873–877. doi: 10.1007/S13361-015-1101-6/ASSET/IMAGES/LARGE/JS8B05035_0002.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garg N., Zeng Y., Edlund A., Melnik A.V., Sanchez L.M., Mohimani H., Gurevich A., Miao V., Schiffler S., Lim Y.W., Luzzatto-Knaan T., Cai S., Rohwer F., Pevzner P.A., Cichewicz R.H., Alexandrov T., Dorrestein P.C. Spatial Molecular Architecture of the Microbial Community of a Peltigera Lichen. mSystems 2016,;1(6) doi: 10.1128/MSYSTEMS.00139-16/ASSET/E213F968-EAE8-4373-8686-29846ACBF1BC/ASSETS/GRAPHIC/SYS0061620740008.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCaughey C.S., Trebino M.A., Yildiz F.H., Sanchez L.M. Utilizing imaging mass spectrometry to analyze microbial biofilm chemical responses to exogenous compounds. Methods Enzymol. 2021;665:281–304. doi: 10.1016/BS.MIE.2021.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B., Dunham S.J.B., Ellis J.F., Lange J.D., Smith J.R., Yang N., King T.L., Amaya K.R., Arnett C.M., Sweedler J.V. A versatile strategy for characterization and imaging of drip flow microbial biofilms. Anal Chem. 2018;90(11):6725–6734. doi: 10.1021/ACS.ANALCHEM.8B00560/ASSET/IMAGES/LARGE/AC-2018-00560J_0005.JPEG. [DOI] [PubMed] [Google Scholar]
- 47.Brockmann E.U., Steil D., Bauwens A., Soltwisch J., Dreisewerd K. Advanced methods for MALDI-MS imaging of the chemical communication in microbial communities. Anal Chem. 2019;91(23):15081–15089. doi: 10.1021/acs.analchem.9b03772. [DOI] [PubMed] [Google Scholar]
- 48.Wakeman C.A., Moore J.L., Noto M.J., Zhang Y., Singleton M.D., Prentice B.M., Gilston B.A., Doster R.S., Gaddy J.A., Chazin W.J., Caprioli R.M., Skaar E.P. The innate immune protein calprotectin promotes pseudomonas aeruginosa and staphylococcus aureus interaction. Nat Commun. 2016;7(1):1–12. doi: 10.1038/ncomms11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prentice B.M., Ryan D.J., Grove K.J., Cornett D.S., Caprioli R.M., Spraggins J.M. Dynamic range expansion by gas-phase ion fractionation and enrichment for imaging mass spectrometry. Anal Chem. 2020;92(19):13092–13100. doi: 10.1021/ACS.ANALCHEM.0C02121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soltwisch J., Kettling H., Vens-Cappell S., Wiegelmann M., Müthing J., Dreisewerd K. Mass spectrometry imaging with laser-induced postionization. Science. 2015;348(6231):211–215. doi: 10.1126/science.aaa1051. [DOI] [PubMed] [Google Scholar]
- 51.Spivey E.C., McMillen J.C., Ryan D.J., Spraggins J.M., Caprioli R.M. Combining MALDI-2 and transmission geometry laser optics to achieve high sensitivity for ultra-high spatial resolution surface analysis. J. Mass Spectrom. 2019;54(4):366–370. doi: 10.1002/jms.4335. [DOI] [PubMed] [Google Scholar]
- 52.Spraggins J.M., Djambazova K.V., Rivera E.S., Migas L.G., Neumann E.K., Fuetterer A., Suetering J., Goedecke N., Ly A., Van de Plas R., Caprioli R.M. High-performance molecular imaging with MALDI trapped ion-mobility time-of-flight (TimsTOF) mass spectrometry. Anal Chem. 2019;91(22):14552–14560. doi: 10.1021/acs.analchem.9b03612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Towers M.W., Karancsi T., Jones E.A., Pringle S.D., Claude E. Optimised desorption electrospray ionisation mass spectrometry imaging (DESI-MSI) for the analysis of proteins/peptides directly from tissue sections on a travelling wave ion mobility Q-ToF. J Am Soc Mass Spectrom. 2018;29(12):2456–2466. doi: 10.1007/s13361-018-2049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barré F., Rocha B., Dewez F., Towers M., Murray P., Claude E., Cillero-Pastor B., Heeren R., Porta Siegel T. Faster raster matrix-assisted laser desorption/ionization mass spectrometry imaging of lipids at high lateral resolution. Int J Mass Spectrom. 2019;437:38–48. doi: 10.1016/j.ijms.2018.09.015. [DOI] [Google Scholar]
- 57.Fernandez-Lima F., Kaplan D.A., Suetering J., Park M.A. Gas-phase separation using a trapped ion mobility spectrometer. International Journal for Ion Mobility Spectrometry. 2011;14(2):93–98. doi: 10.1007/s12127-011-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez-Lima F.A., Kaplan D.A., Park M.A. Note: integration of trapped ion mobility spectrometry with mass spectrometry. Rev. Sci. Instrum. 2011;82(12) doi: 10.1063/1.3665933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jackson S.N., Barbacci D., Egan T., Lewis E.K., Albert Schultz J., Woods A.S. MALDI-ion mobility mass spectrometry of lipids in negative ion mode. Anal. Methods. 2014;6(14):5001–5007. doi: 10.1039/c4ay00320a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLean J.A., Ridenour W.B., Caprioli R.M. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J. Mass Spectrom. 2007;42(8):1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 61.Feider C.L., Elizondo N., Eberlin L.S. Ambient ionization and FAIMS mass spectrometry for enhanced imaging of multiply charged molecular ions in biological tissues. Anal Chem. 2016;88(23):11533–11541. doi: 10.1021/acs.analchem.6b02798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Djambazova, K.; Klein, D. R.; Migas, L.; Neumann, E.; Rivera, E.; Van de Plas, R.; Caprioli, R. M.; Spraggins, J. Resolving the Complexity of Spatial Lipidomics with MALDI Trapped Ion Mobility Spectrometry. 2020. https://doi.org/10.26434/CHEMRXIV.12331652.V1. [DOI] [PubMed]
- 63.Brindhadevi K., LewisOscar F., Mylonakis E., Shanmugam S., Verma T.N., Pugazhendhi A. Biofilm and quorum sensing mediated pathogenicity in pseudomonas aeruginosa. Process Biochem.. Elsevier Ltd September 1. 2020:49–57. doi: 10.1016/j.procbio.2020.06.001. [DOI] [Google Scholar]
- 64.Nelson K.A., Daniels G.J., Fournie J.W., Hemmer M.J. Optimization of whole-body zebrafish sectioning methods for mass spectrometry imaging. J Biomol Tech. 2013;24(3):119. doi: 10.7171/JBT.13-2403-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H.Y.J., Liu C. A simple desalting method for direct MALDI mass spectrometry profiling of tissue lipids. J Lipid Res. 2011;52(4):840–849. doi: 10.1194/jlr.D013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moormeier D.E., Bayles K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol Microbiol. 2017;104(3):365–376. doi: 10.1111/MMI.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bleich R., Watrous J.D., Dorrestein P.C., Bowers A.A., Shank E.A. Thiopeptide antibiotics stimulate biofilm formation in bacillus subtilis. Proc Natl Acad Sci U S A. 2015;112(10):3086–3091. doi: 10.1073/PNAS.1414272112/SUPPL_FILE/PNAS.201414272SI.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koprivnjak T., Zhang D., Ernst C.M., Pesche A., Nauseef W.M., Weiss J.P. Characterization of staphylococcus aureus cardiolipin synthases 1 and 2 and their contribution to accumulation of cardiolipin in stationary phase and within phagocytes. J Bacteriol. 2011;193(16):4134–4142. doi: 10.1128/JB.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Short S.A., White D.C. Metabolism of phosphatidylglycerol, lysylphosphatidylglycerol, and cardiolipin of staphylococcus aureus. J Bacteriol. 1971;108(1):219–226. doi: 10.1128/jb.108.1.219-226.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Missiakas, D. M.; Schneewind, O. Growth and Laboratory Maintenance of Staphylococcus Aureus. Curr Protoc Microbiol2013, 28 (1), 9C.1.1-9C.1.9. https://doi.org/10.1002/9780471729259.MC09C01S28. [DOI] [PMC free article] [PubMed]
- 71.Kohler C., Proctor R.A., Bayer A.S., Yeaman M.R., Lalk M., Engelmann S., Mishra N.N. Proteomic and membrane lipid correlates of re-duced host defense peptide susceptibility in a snod mutant of staphylococcus aureus. Antibiotics. 2019;8(4) doi: 10.3390/ANTIBIOTICS8040169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yarwood J.M., Bartels D.J., Volper E.M., Greenberg E.P. Quorum sensing in staphylococcus aureus biofilms. J Bacteriol. 2004;186(6):1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kennedy A.D., Otto M., Braughton K.R., Whitney A.R., Chen L., Mathema B., Mediavilla J.R., Byrne K.A., Parkins L.D., Tenover F.C., Kreiswirth B.N., Musser J.M., DeLeo F.R. Epidemic community-associated methicillin-resistant staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A. 2008;105(4):1327–1332. doi: 10.1073/PNAS.0710217105/SUPPL_FILE/10217TABLE7.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ryan D.J., Patterson N.H., Putnam N.E., Wilde A.D., Weiss A., Perry W.J., Cassat J.E., Skaar E.P., Caprioli R.M., Spraggins J.M. MicroLESA: integrating autofluorescence microscopy, in situ micro-digestions, and liquid extraction surface analysis for high spatial resolution targeted proteomic studies. Anal Chem. 2019;91(12):7578–7585. doi: 10.1021/ACS.ANALCHEM.8B05889/ASSET/IMAGES/LARGE/AC-2018-058898_0004.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS Online Tools for Lipid Research. Nucleic Acids Res2007, 35 (suppl_2), W606–W612. https://doi.org/10.1093/NAR/GKM324. [DOI] [PMC free article] [PubMed]
- 76.Fahy E., Subramaniam S., Murphy R.C., Nishijima M., Raetz C.R.H., Shimizu T., Spener F., van Meer G., Wakelam M.J.O., Dennis E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50:S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.