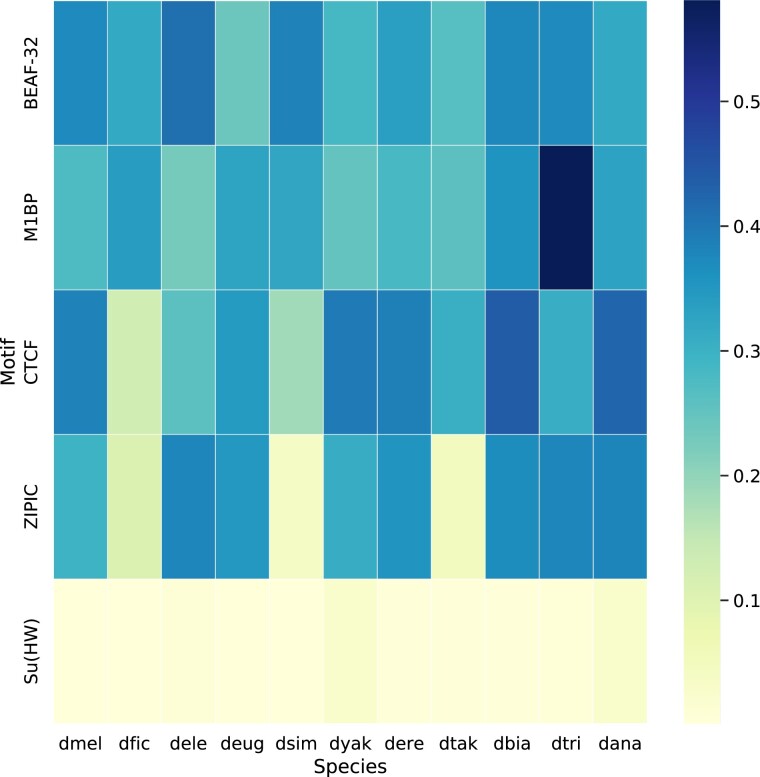
Fig. 2.
Boundary motif enrichment. We used AME (v. 5.3.3) from MEME suite (Bailey et al. 2009) to measure the enrichment of sequence motifs for five insulator proteins whose binding sites have previously been shown to be associated with TAD boundaries in D. melanogaster. Shown here are the percentage of boundaries containing at least one sequence motif (a value reported as %TP by AME). The heatmap shows %TP for the BEAF-32, M1BP, CTCF, ZIPIC, and Su(HW) insulator protein binding motifs at the boundaries of all 11 species used in this study. We found consistent enrichment of M1BP and BEAF-32 at TAD boundaries across all species, both of which are known to be important for TAD boundary formation in D. melanogaster (Ramírez et al. 2018). CTCF, ZIPIC, and Su(HW) motifs are found at a relatively small number of TAD boundaries in D. melanogaster (Ramírez et al. 2018). CTCF and ZIPIC show variable enrichment across species, whereas Su(HW) shows consistently weak enrichment.