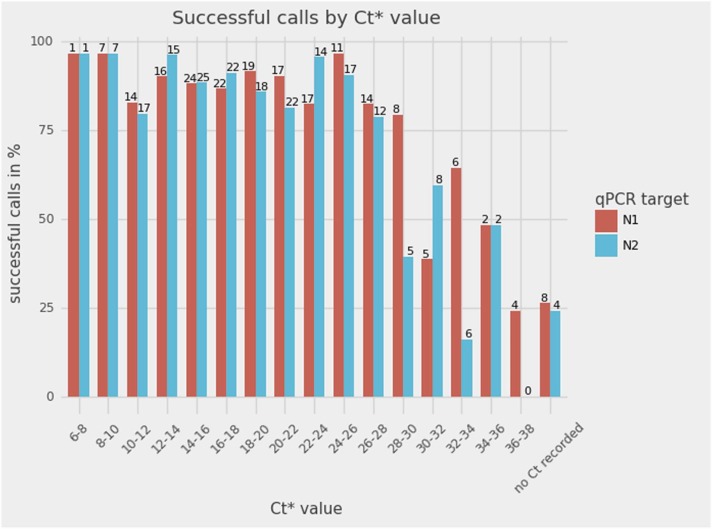
Fig. 3.
The sensitivity of the SpikeSeq method versus the Ct values of the diagnostic RT-qPCR test (n = 195). The number above each bar refers to the number of samples in that bin. Overall, Sanger sequencing data from 85% of samples were of sufficient quality for mapping. The non-template control is not depicted, as it did not produce a Sanger read that mapped to the reference genome. * : Ct values stem from a 2-step PCR where the first 7 cycles are not fluorescence registered, possibly meaning that the Sanger sequencing assay is even more sensitive than suggested by this figure.