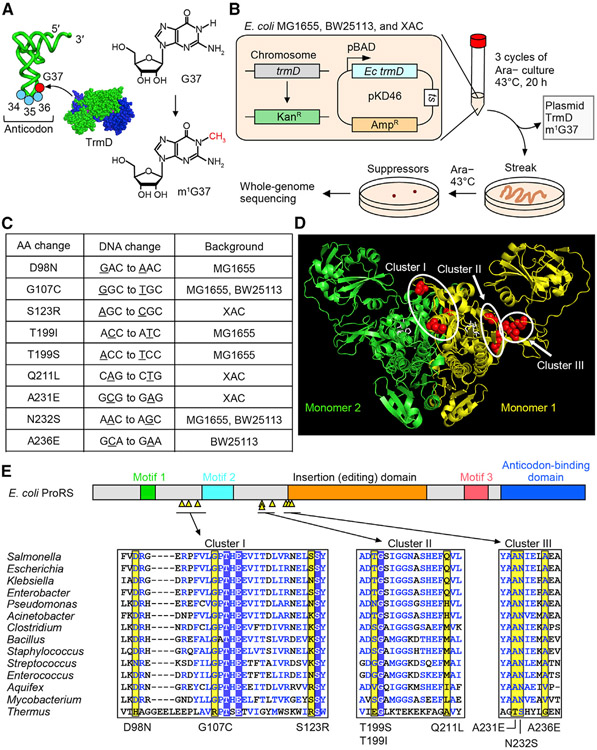
Figure 1. E. coli trmD-KO suppressors are mapped to proS.
(A) Synthesis of m1G37 (red) by TrmD on the 3′ side of the anticodon at positions 34–36 (blue).
(B) E. coli trmD-KO in MG1655, BW25113, and XAC for isolation of suppressors.
(C) Exclusive mapping of all suppressor mutations to proS (Table S1).
(D) Clusters of suppressor mutations in the crystal structure of the Enterococcus faecalis proS enzyme with a prolyl analog (PDB: 2J3L). Mutations are mapped to residues in red in monomer #1, while the prolyl analog is in white.
(E) (Top) Suppressor mutations (yellow triangles) in a linear diagram of E. coli proS structural domains. (Bottom) A multi-sequence alignment of the enzyme, showing residues with mutations (yellow), those conserved (boxed in blue), and those in structural similarity (blue letters). An expanded alignment is in Figure S2C.