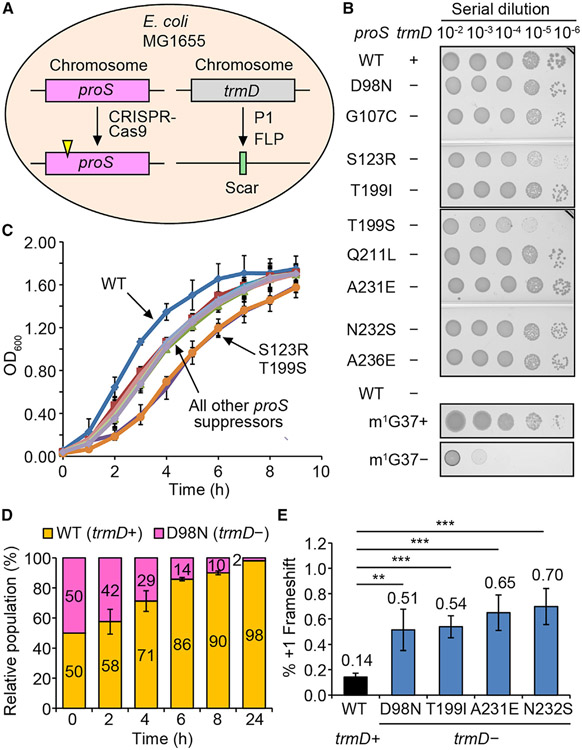
Figure 2. Each proS suppressor supports cell viability without trmD.
(A) CRISPR-Cas9 reconstruction of each suppressor in MG1655.
(B) A serial dilution of each reconstructed suppressor on an Luria-Bertani (LB) plate at 37°C. WT is the native MG1655, while each suppressor is shown by the proS mutation. The plus (+) and minus (−) symbols indicate the presence and absence of the chromosomal trmD. Growth of MG1655-trmD-KO is shown at the bottom with or without Ara, representing m1G37+ and m1G37− conditions.
(C) Growth of each reconstructed suppressor at 37°C. Data are mean ± SD (n = 3).
(D) A 1:1 mixture of the WT (expressing YFP) and the proS-D98N suppressor (expressing mCherry) monitored for cell fitness at 37°C in LB. Data are mean ± SD (n = 3).
(E) A +1-frameshifting assay for expression of nLuc in MG1655 or in a reconstructed suppressor. Data are mean ± SD (n = 4). **p < 0.05, ***p < 0.01.