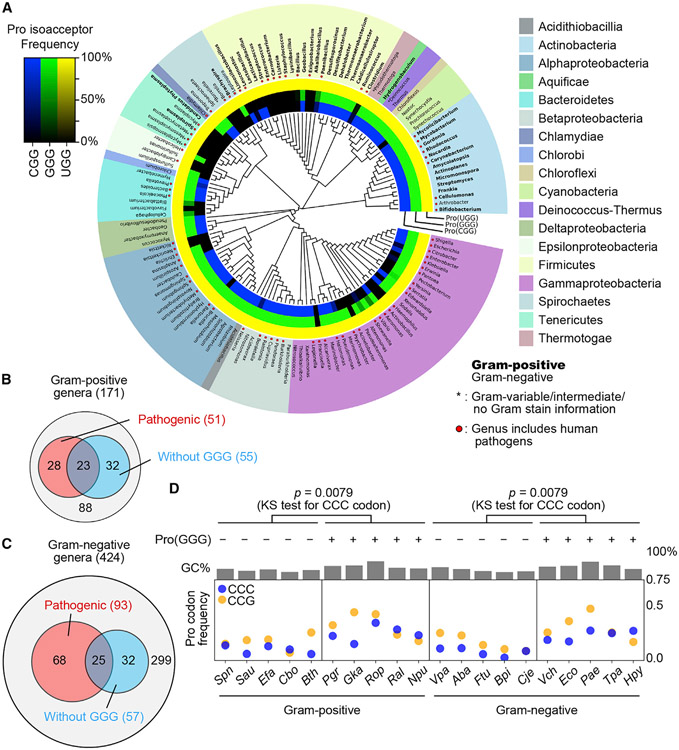
Figure 6. Absence of Pro(GGG) from some bacterial species.
(A) A phylogenetic tree showing the presence and absence of tRNAPro genes in representative bacterial genomes at GtRNAdb. Gram-positive (Gram (+)) genera are in bold face, and those of human pathogens are indicated by a red dot. Inner rings show the presence and absence of tRNAPro genes: blue for Pro(CGG), green for Pro(GGG), and yellow for Pro(UGG). The gradient of color darkness indicates the frequency of the species lacking the corresponding tRNA gene in the genus. Darker color means more species lacking the tRNA gene, with the black indicating absence.
(B) Distribution of Gram-positive genera of medical relevance without Pro(GGG).
(C) Distribution of Gram-negative genera of medical relevance without Pro(GGG).
(D) Average frequency of CCC and CCG codons in each protein-coding gene across gram-positive and -negative bacteria with or without Pro(GGG). The p value is for analysis of CCC. KS test, Kolmogorov-Smirnov test. Species names are in STAR Methods.