Figure 1. Study Design.
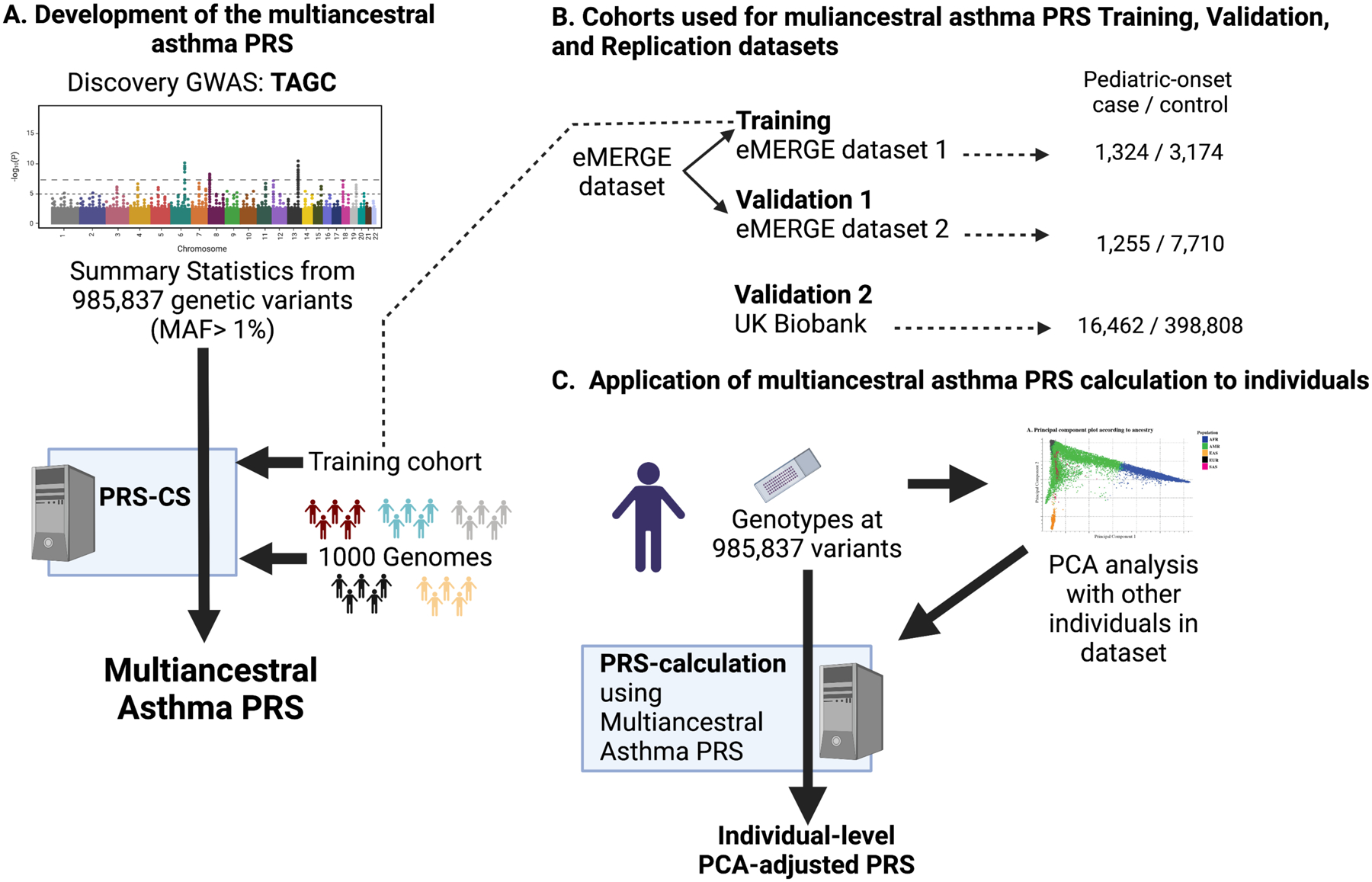
(A) Summary statistics from 985,837 genetic variants with minor allele frequencies (MAF) greater than 1% in the overall (cases and controls combined) Trans-National Asthma Genetic Consortium (TAGC) Discovery genome-wide association studies (GWAS) were used to develop the multiancestral asthma PRS in the context of the linkage disequilibrium of the reference 1000 genomes reference panel and the Training dataset. (B) The eMERGE dataset was split into two independent datasets (Training and Validation 1) and the UK Biobank was used as a Validation 2 dataset. Subjects with confirmed pediatric-onset asthma and controls were used for PRS Training, Validation 1, and Validation 2. C) Individual genotypes from each subject in each dataset was used to perform a principal component analysis (PCA). For each individual, genotypes at each of the 985,837 variants included in the multiancestral asthma PRS were used to calculate a PCA-adjusted PRS. The adjusted multiancestral PRS was applied to each individual in the Training, Validation 1, and Validation 2 cohorts in preparation for it to be similarly applied to individuals recruited into an IRB-approved prospective intervention study. Figure created in BioRender.
