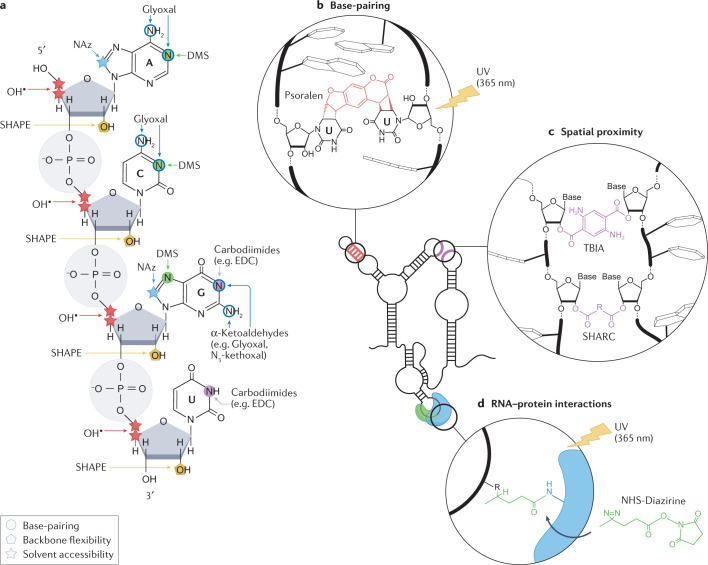
Fig. 1. Chemical probes for interrogating RNA structures.
a, Targets of different chemical probes on RNA, including dimethyl sulfate (DMS), α-ketoaldehydes (such as Glyoxal and N3-kethoxal), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), selective 2′-hydroxyl acylation analysed by primer extension (SHAPE) reagents, hydroxyl radicals and nicotinoyl azide (NAz). Sites of chemical modification by probes measuring the pairing status of nucleobases (circles), the solvent accessibility of RNA residues (stars) and the flexibility of the RNA backbone (pentagons) are marked. b, Psoralen interacts with uridines on opposite strands of an RNA duplex and mediates cross-linking of the two strands upon long-wave UV irradiation (365 nm). Cross-linking can occur both intramolecularly and intermolecularly. c, The reaction of bifunctional acylating compounds, such as trans-bis-isatoic anhydride (TBIA) and spatial 2′-hydroxyl acylation reversible cross-linking (SHARC) reagents, results in cross-links between structurally flexible nucleotides that are spatially proximal to each other. Cross-linking can occur both intramolecularly and intermolecularly. d, Upon long-wave UV irradiation, NHS-diazirine cross-links RNA nucleotides and amino acids (usually lysine) of interacting proteins at the RNA–protein interaction interface.