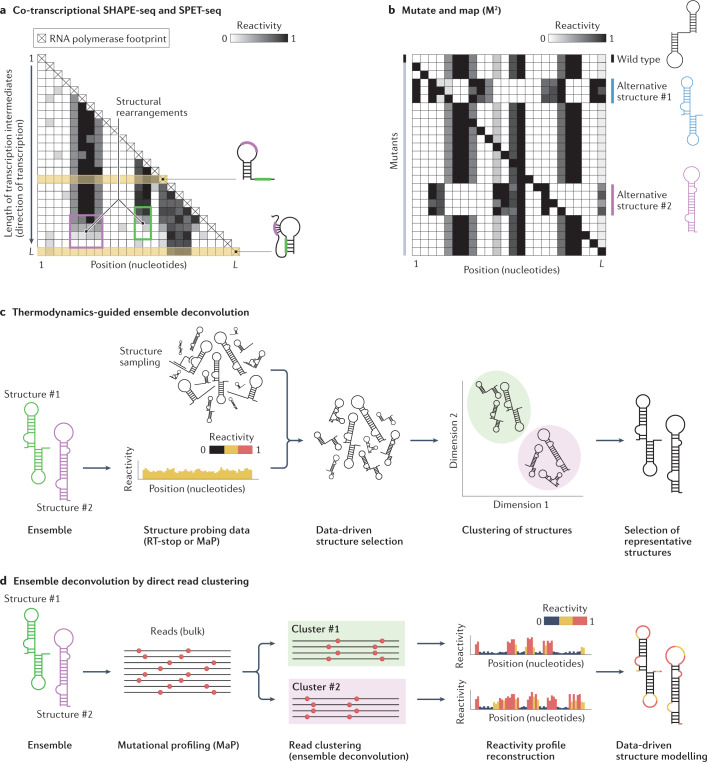
Fig. 5. Experimental and computational methods for RNA ensemble deconvolution.
a, Assays such as co-transcriptional selective 2′-hydroxyl acylation analysed by primer extension (SHAPE) followed by sequencing (SHAPE-seq) and structural probing of elongating transcripts followed by sequencing (SPET-seq) allow RNA co-transcriptional structure folding pathways to be deconvolved by first probing the entire population of transcription intermediates, followed by the computational reconstruction of the individual reactivity profiles. Plotting these reactivity profiles in the form of a heatmap, with the rows corresponding to distinct transcription intermediates sorted by increasing length, provides intuitive visualization of RNA structural rearrangements occurring as transcription proceeds (top to bottom). The example shows two transcription intermediates, each represented by the rows denoted in yellow. During the transition from the first to the second intermediate, the reactivity of the unpaired regions (coloured purple and green on the structures) progressively drops (purple and green boxes on the heatmap) as they begin to undergo base-pairing, resulting in a pseudoknot (purple region) and a stem-loop (SL) (green region). b, Mutate and map (M2) provides an indirect way to deconvolve RNA structure ensembles by randomly generating a large number of single-nucleotide substitution mutants of an RNA of interest, followed by structure probing analysis. Mutations capable of disrupting base-pairing interactions in the wild-type structure, whilst stabilizing alternative folds, will cause a redistribution of the relative abundance of the structures within the ensemble, leading to reactivity changes. The reactivity profiles of these mutants can then be used to infer the structure of these alternative conformations. c, The first group of computational methods for ensemble deconvolution exploits thermodynamics-guided RNA structure prediction software to sample a large number of structures from the theoretical ensemble the RNA of interest can form, and then uses the experimental data to select the smallest possible subset of structures that can explain the data. Typically, structures are then clustered together by similarity and a single representative structure is returned for each cluster. This class of approaches is suitable for the analysis of both reverse transcriptase (RT)-stop and mutational profiling (MaP) RNA structure probing data. d, The second group of computational methods for ensemble deconvolution involves direct read clustering. These methods take sequencing reads from MaP experiments and attempt to define clusters of reads with correlated patterns of mutations, corresponding to alternative RNA conformations. Clustered reads can be processed into reactivity profiles that can then be used to inform structure modelling.