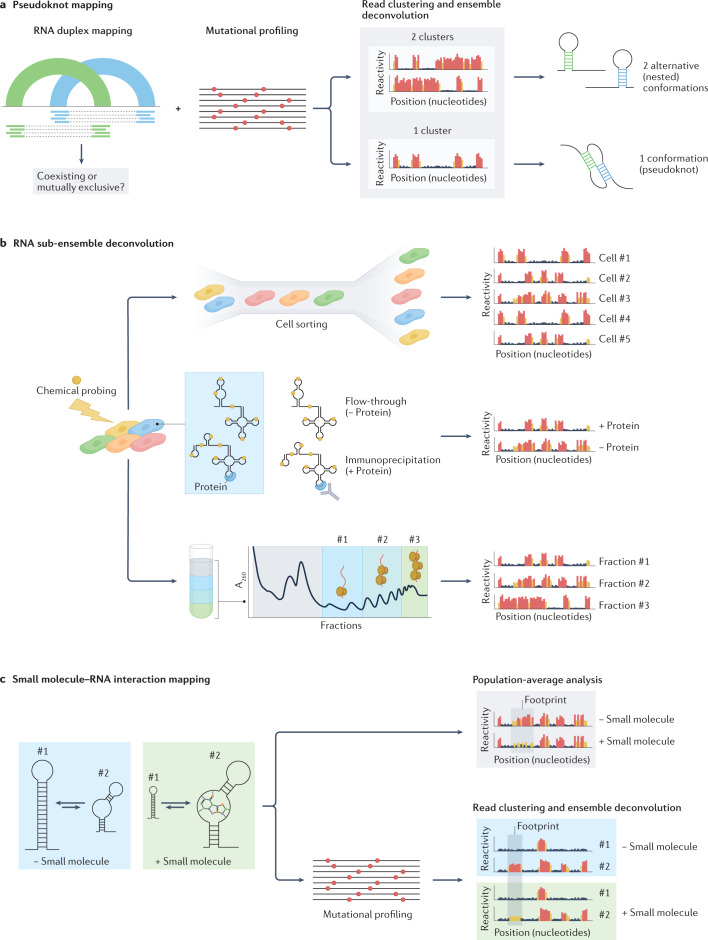
Fig. 7. Challenges in high-throughput sequencing (HTS)-based RNA structure mapping studies.
a, Mapping of pseudoknots can potentially be achieved by combining direct RNA–RNA interaction capture with methods for ensemble deconvolution from chemical probing experiments. Although RNA duplex mapping does not preserve any information about the relationship between two independent helices, using ensemble deconvolution analysis to determine whether the region of the RNA encompassing these helices populates one or two conformations can help determine whether two incompatible helices coexist within the same RNA molecule, forming a pseudoknot, or whether they belong to two independent RNA molecules. b, Specialized structure probing assays can aid the analysis of RNA structure ensembles in vivo. Coupling of chemical probing with single-cell analysis (top), RNA immunoprecipitation (middle) or polysome fractionation (bottom) would increase the resolution of RNA structure analyses, possibly enabling the characterization of lowly abundant RNA conformations. c, RNA chemical probing can aid the mapping of small molecule–RNA interactions. Analysis of population-averaged reactivities can be used to identify footprints of small molecules binding to RNA. The coupling of chemical probing with ensemble deconvolution analysis can further help elucidate binding modes of small molecules, possibly enabling the identification of specific RNA conformations targeted by the small molecule.