Abstract
Grapevine downy mildew is one of the most devastating diseases in grape production worldwide, but its pathogenesis remains largely unknown. A thorough understanding of the interaction between grapevine and the causal agent, Plasmopara viticola, is helpful to develop alternative disease control measures. Effector proteins that could be secreted to the interaction interface by pathogens are responsible for the susceptibility of host plants. In this study, a Crinkler effector, named PvCRN17, which is from P. viticola and showed virulent effects towards Nicotiana benthamiana previously, was further investigated. Consistently, PvCRN17 showed a virulent effect on grapevine plants. Protein–protein interaction experiments identified grapevine VAE7L1 (Vitis protein ASYMMETRIC LEAVES 1/2 ENHANCER 7‐Like 1) as one target of PvCRN17. VAE7L1 was found to interact with VvCIA1 and VvAE7, thus it may function in the cytosolic iron–sulphur cluster assembly (CIA) pathway. Transient expression of VAE7L1 in Vitis riparia and N. benthamiana leaves enhanced the host resistance to oomycete pathogens. Downstream of the CIA pathway in grapevine, three iron–sulphur (Fe‐S) proteins showed an enhancing effect on the disease resistance of N. benthamiana. Competitive co‐immunoprecipitation assay showed PvCRN17 could compete with VvCIA1 to bind with VAE7L1 and VvAE7. Moreover, PvCRN17 and VAE7L1 were colocalized at the plasma membrane of the plant cell. To conclude, after intruding into the grapevine cell, PvCRN17 would compete with VCIA1 to bind with VAE7L1 and VAE7, demolishing the CIA Fe‐S cluster transfer complex, interrupting the maturation of Fe‐S proteins, to suppress Fe‐S proteins‐mediated defence responses.
Keywords: CIA pathway, CRN effector, Fe‐S proteins, grapevine, Plasmopara viticola, virulence
Two indispensable components of the cytosolic iron–sulphur cluster assembly pathway in grapevine are abducted to the plasma membrane by a virulent CRN effector from Plasmopara viticola to impede grapevine defence responses.
1. INTRODUCTION
Resolving the causes of crop diseases is pivotal to the agricultural industry. Efficient control measures developed according to the pathogenesis of a certain crop disease would help stopping the spread of the disease, reducing or avoiding quality and yield loss of agricultural products. Plants showing different degrees of resistance to a certain disease results from the complicated interactions between themselves and the pathogen, which usually belongs to bacteria, fungi, oomycetes or nematodes (Dangl et al., 2013; Jones & Dangl, 2006; Miller et al., 2017). The mechanisms of plant–pathogen interactions have been revealed from a macroscopic to a microscopic level by human endeavour. In the molecular aspect, the rough view of plant–pathogen interactions is described as follows: there are proteins with the leucine rich‐repeat (LRR) domain on the plant cell surface, which act as receptors for nonself‐recognition or danger sensing. Once the pathogen arrives at the plant cell surface after crossing over the plant physical barriers (Łaźniewska et al., 2012), its conserved molecular structures such as flagellin, peptidoglycans, chitins and β‐glucans, will bind to the cytoplasmic membrane localized LRR receptor proteins (Nürnberger & Kemmerling, 2009). Following the binding, the signals of nonself‐invasion are received by the plant cell. Then, a series of defence responses is triggered, including calcium influx, production of reactive oxygen species (ROS), activation of MITOGEN‐ACTIVATED PROTEIN KINASES (MAPKs), increased biosynthesis of salicylic acid (SA), and up‐regulation of defence‐related genes (Peng et al., 2018; Spoel & Dong, 2012; Tian et al., 2021). Responses related to these early reactions of the plant cell are collectively named pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI). However, substances for delaying, interfering with, or blocking up the defence responses, including PTI, are often secreted by the pathogen onto the interaction interface to promote infection. For example, pathogens produce secretory proteins called effectors that are delivered to the apoplast or into the plant cell, promoting their virulence. Among the cytoplasmic effectors, Crinkler (Crinkling and necrosis proteins, CRN) and RXLR (possessing the conserved arginine‐any amino acid‐leucine‐arginine motif) effectors are the most notorious molecules secreted by various pathogens to restrain the immune responses of host plants (Kamoun, 2006; Schornack et al., 2009). Concomitantly, there are nucleotide‐binding, LRR receptors (NLRs) within the plant cell that can interact with some effectors directly or indirectly. On recognition of the effectors by NLR proteins, stronger defence responses, collectively called effector‐triggered immunity (ETI), are elicited, protecting the plant from further damage (Dangl et al., 2013; Song et al., 2021). ETI uses signalling components involved in PTI, and there are convergent signalling components between the processes of PTI and ETI as well (Ngou et al., 2022; Peng et al., 2018; Pruitt et al., 2021; Yuan et al., 2021). Generally, plants struggle to survive with PTI and ETI, while pathogens fight to parasitize their hosts by delivering effectors into the interaction interface.
In addition to the defence reactions mentioned above, there are various substances integrated into the sophisticated plant immune system (Miller et al., 2017). Recently, several plant iron–sulphur proteins and related iron–sulphur cluster assembly systems have been reported to be involved in plant–pathogen interactions. For example, ferredoxin‐like proteins from Arabidopsis thaliana and Nicotiana benthamiana, which are iron–sulphur proteins, have been shown to be indispensable for host and nonhost resistance to microbes (Fonseca et al., 2020, 2021; Moeder et al., 2007; Wang et al., 2018).
Iron–sulphur proteins (Fe‐S proteins) are proteins incorporated with inorganic iron–sulphur clusters ([Fe2S2], [Fe4S4], or [Fe3S4]) and commonly produced in organisms from prokaryotes to eukaryotes. Studies have revealed that Fe‐S proteins are related to electron transfer and energy transfer (such as respiration and photosynthesis), redox reaction, gene expression regulating, DNA replication and repair, genome integrity maintenance, and protein translation (Balk & Lobréaux, 2005; Balk & Schaedler, 2014; Couturier et al., 2015; Johnson et al., 2005; Lill, 2009; Lill & Mühlenhoff, 2008; Rouault, 2019; Tong & Rouault, 2007). Mature Fe‐S proteins are formed by the assembling of Fe‐S clusters into the precursors of Fe‐S proteins (apoproteins) by specific mechanisms. For plants, there are three iron–sulphur cluster assembly pathways for the maturation of different Fe‐S proteins: the sulphur mobilization (SUF)‐like machinery in the plastids, the iron–sulphur cluster (ISC) pathway in the mitochondria, and the cytosolic iron–sulphur cluster assembly (CIA) pathway in the cytosol. The maturation of Fe‐S proteins localized in the cytosol and nuclei depends on both the ISC and CIA pathways (Balk & Pilon, 2011; Balk & Schaedler, 2014). Defects in the components of the CIA pathway, such as CIA1, AE7 and MET18 in A. thaliana, cause dysfunction of some Fe‐S proteins, such as the tricarboxylic acid cycle‐related aconitases and the DNA glycosylase DEMETER‐Like 1 (ROS1), resulting in DNA damage, whole‐genome DNA hypermethylation, or growth arrest of the mutant plants (Duan et al., 2015; Luo et al., 2012; Wang et al., 2016). Because some Fe‐S proteins are involved in plant–pathogen interactions, the importance of iron–sulphur cluster assembly pathways to the plant immune system will be discovered in the future. This study reports that a virulent CRN effector from Plasmopara viticola, PvCRN17 (Xiang et al., 2021), could interact with components of the plant CIA pathway, including protein AE7‐like 1 in Vitis genus. The findings could help to determine the molecular mechanism used by P. viticola to promote its infection on the host grapevine.
2. RESULTS
2.1. PvCRN17 shows virulent effects on plants
In agreement with the previous study (Xiang et al., 2021), when transiently expressed in N. benthamiana leaves, PvCRN17 decreased the host resistance to the hemibiotrophic pathogen Phytophthora capsici (Figure 1a,b). H2O2 accumulation in the inoculated leaves expressing PvCRN17 was weaker than that of the control expressing GFP (Figure 1c,d). Previously, PvCRN17 was characterized by the suppressing effect on PTI induced by the elicitor INF1 (Xiang et al., 2021). Here, PvCRN17 could block the cell death in N. benthamiana leaves induced by the RXLR effector PvAvh74, which is also from the P. viticola isolate YL (Yin et al., 2019). However, PvCRN17 failed to completely inhibit plant hypersensitive cell death induced by PvRXLR77 (Figure 1e–h), which shows avirulent effect on N. benthamiana and the resistant grapevine Vitis riparia (Xiang et al., 2021). However, PvCRN17 could impede some ETI induced by certain effectors in plants.
FIGURE 1.
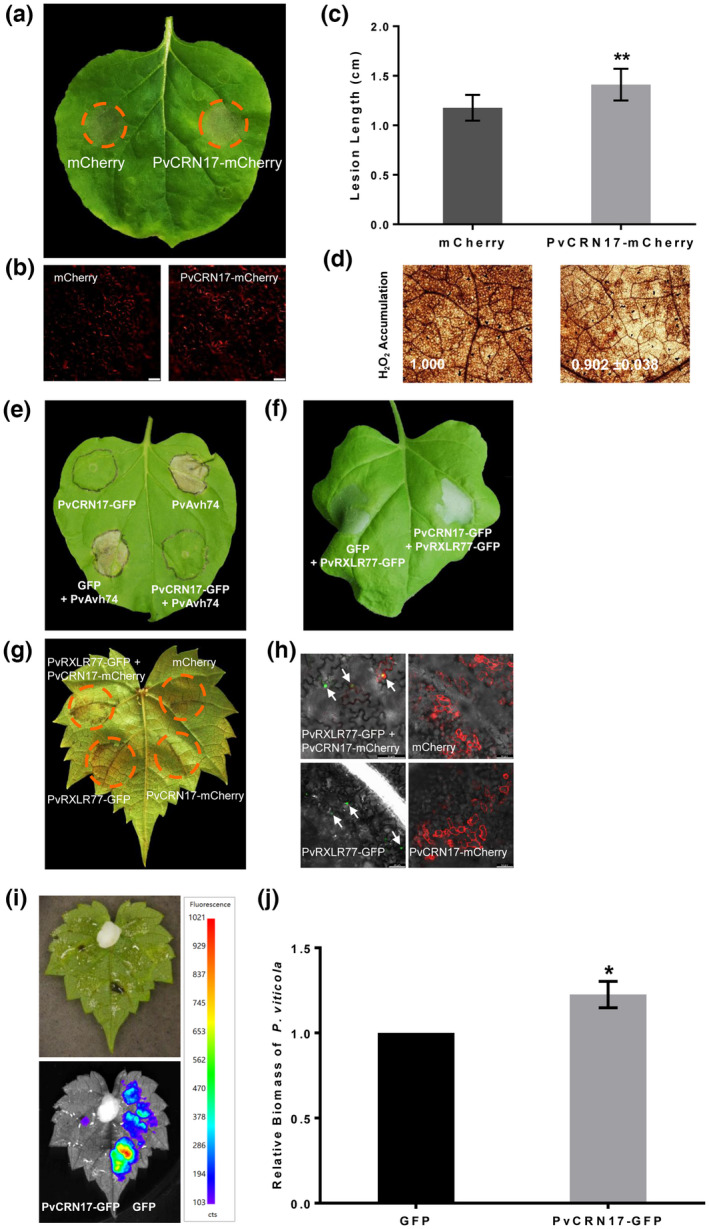
PvCRN17 could suppress reactive oxygen species accumulation in Nicotiana benthamiana leaves challenged with Phytophthora capsici and attenuate the disease resistance of grapevine to Plasmopara viticola. (a) PvCRN17 promoted the susceptibility of N. benthamiana leaves to P. capsici. (b) Confirmation of the fluorescence of mCherry and PvCRN17‐mCherry in N. benthamiana leaves. (c) Mean lesion lengths measured corresponding to the inoculated N. benthamiana leaves. (d) H2O2 accumulation in the inoculation sites were attenuated by PvCRN17. Numbers indicate the relative staining intensity. (e) PvCRN17 suppressed hypersensitive cell death in N. benthamiana leaves induced by PvAvh74. (f) PvCRN17 failed to suppress the hypersensitive cell death in N. benthamiana leaves induced by PvRXLR77. (g) PvCRN17 partially suppressed hypersensitive cell death in Vitis riparia leaves induced by PvRXLR77. (h) Confirmation of the presence of indicated proteins in V. riparia leaves by fluorescence detection. (i) Confirmation of the expression of PvCRN17‐GFP and GFP in V. riparia leaves. (j) PvCRN17 promoted P. viticola colonization on V. riparia leaves when transiently expressed. Error bars represent the standard deviation (SD). Asterisks indicate significant differences from the control groups (paired t test; *p < 0.05, **p < 0.01). The arrowheads denote the nucleus, which is the main destination of PvRXLR77‐GFP. Similar results were obtained in three independent experiments
As a foreign protein to grapevine under natural conditions, the effect of PvCRN17 on the disease resistance of grapevine was tested via transient expression. In vitro V. riparia leaves were infiltrated with agrobacteria on the right and left regions from the abaxial side to express GFP and PvCRN17‐GFP, respectively. Grapevine leaves expressing GFP and PvCRN17‐GFP were both inoculated with equivalent P. viticola sporangial suspension, then the disease severity was estimated by the relative biomass of the pathogen colonizing the leaves. The average relative biomass of P. viticola parasitizing the leaves expressing PvCRN17‐GFP was significantly higher than that of the control (Figure 1i,j), which suggests that PvCRN17 has a virulent effect on grapevine.
To further validate the function of PvCRN17 in the host plant, transgenic Arabidopsis plants transformed with PvCRN17‐mCherry were obtained. Transgenic T1 plants containing PvCRN17‐mCherry (line 1 to line 8) all showed apparent growth arrest compared to the wild‐type plants (WT); for example, there were fewer leaves and flowers on the transgenic plants than on the WT (Figure S1). Transgenic plant line 8 did not live long enough to flower and line 7 failed to generate ripe siliques. Meanwhile, T2 generations of the other transgenic lines failed to germinate on the selection medium (data not shown). These phenomena indicate that PvCRN17 might interfere with certain cellular process of the host, leading to defects in plant development.
It is thought that the carboxyl‐terminus of a CRN effector determines the virulence of the whole molecule (Haas et al., 2009; Schornack et al., 2010). A predicted transmembrane helix structure (TMH) was found at the C‐terminal region (144–163) of PvCRN17 (Xiang et al., 2021). Correspondingly, this study found that the recombinant protein PvCRN17‐GFP could not be detected by western blot unless sodium deoxycholate was added to the plant total protein extraction buffer (Figure 2a). Sodium deoxycholate is effective and necessary for the extraction of transmembrane proteins; therefore, with its TMH, PvCRN17 could be tightly embedded into the plant plasma membrane. Furthermore, when transiently expressed in N. benthamiana leaves and grapevine leaves, a mutant of PvCRN17, PvCRN17‐tmh, with the TMH‐related motif mutated (VLMFG to KKKKK), was mainly distributed in the cytoplasm of the plant cell (Figure 2b,c). Compared with PvCRN17, PvCRN17‐tmh showed compromised suppression activity on the plant cell death induced by Bax (Figure 2d). Moreover, the plant cell death induced by PvAvh74 could not be impeded by PvCRN17‐tmh (Figure 2e). Therefore, PvCRN17 mainly occupies the plasma membrane of the host plant cell and its destination contributes to its virulence.
FIGURE 2.
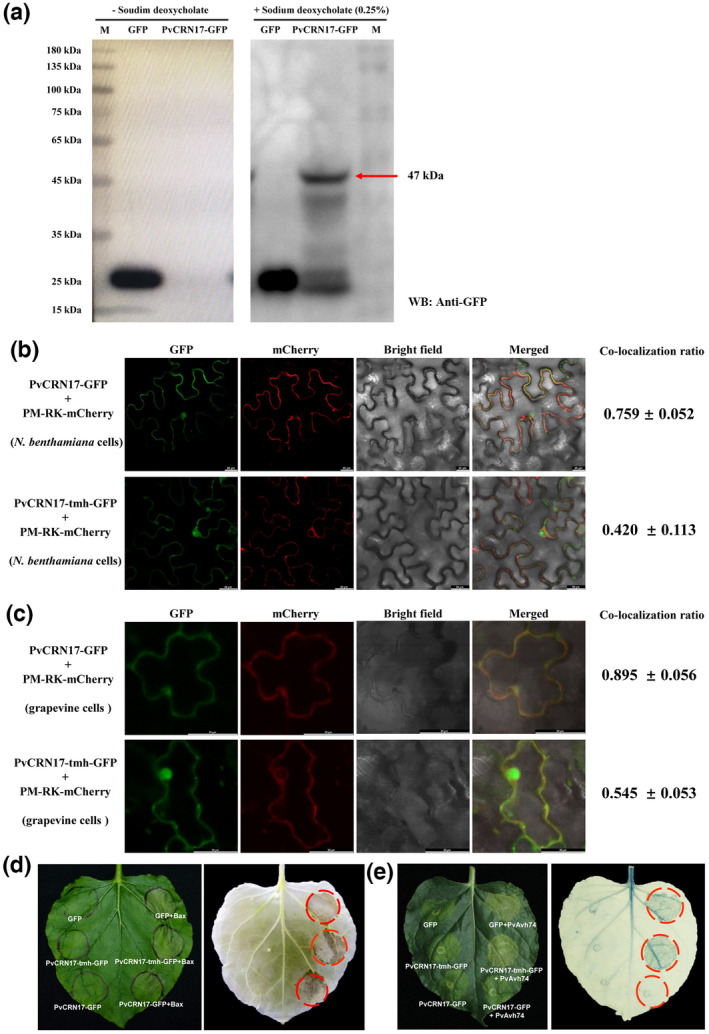
PvCRN17 is mainly localized in the plasma membrane of the plant cell and its virulence depends on its destination. (a) PvCRN17‐GFP expressed in Nicotiana benthamiana leaves could be fully extracted only after adding sodium deoxycholate into the extraction buffer, otherwise the recombinant protein (molecular weight 47 kDa) could not be detected by western blotting. “− Sodium deoxycholate” means no sodium deoxycholate in the protein extraction buffer, “+ Sodium deoxycholate (0.25%)” means that 0.25% sodium deoxycholate was added. (b, c) PvCRN17‐tmh with the transmembrane helix in PvCRN17 mutated was mainly distributed in the cytoplasm of the plant cell when transiently expressed in N. benthamiana and Vitis riparia leaves. (d) The suppression activity of PvCRN17‐tmh on programmed cell death induced by Bax was weaker than that of PvCRN17. (e) PvCRN17‐tmh failed to suppress the cell death induced by PvAvh74 in N. benthamiana leaves compared with PvCRN17
2.2. PvCRN17 interacts with plant AE7‐Like 1 proteins
Finding the targets of PvCRN17 in grapevine is necessary to explain its virulent effect. In this study, a yeast‐two hybrid (Y2H) assay was conducted to identify target proteins in the grapevine of PvCRN17 by screening the mixed cDNA library of Vitis vinifera ‘Pinot Noir’ and Vitis piasezkii ‘Liuba‐8’ leaves challenged with P. viticola. Several candidate prey proteins were screened out (Table S1). Finally, V. vinifera protein AE7‐Like 1 (VvAE7L1) and nitrogen regulatory protein P‐II homologue isoform X1 (NRPPII‐X1) were confirmed to interact with PvCRN17 in yeast (Figure 3a). To confirm the interactions, bimolecular fluorescence complementation (BiFC) and co‐immunoprecipitation (Co‐IP) assay were conducted. In accordance with the Y2H assay, both BiFC and Co‐IP assays supported that PvCRN17 interacted with VvAE7L1 as well as VvNRPPII‐X1 in vivo (Figure 3b–d).
FIGURE 3.
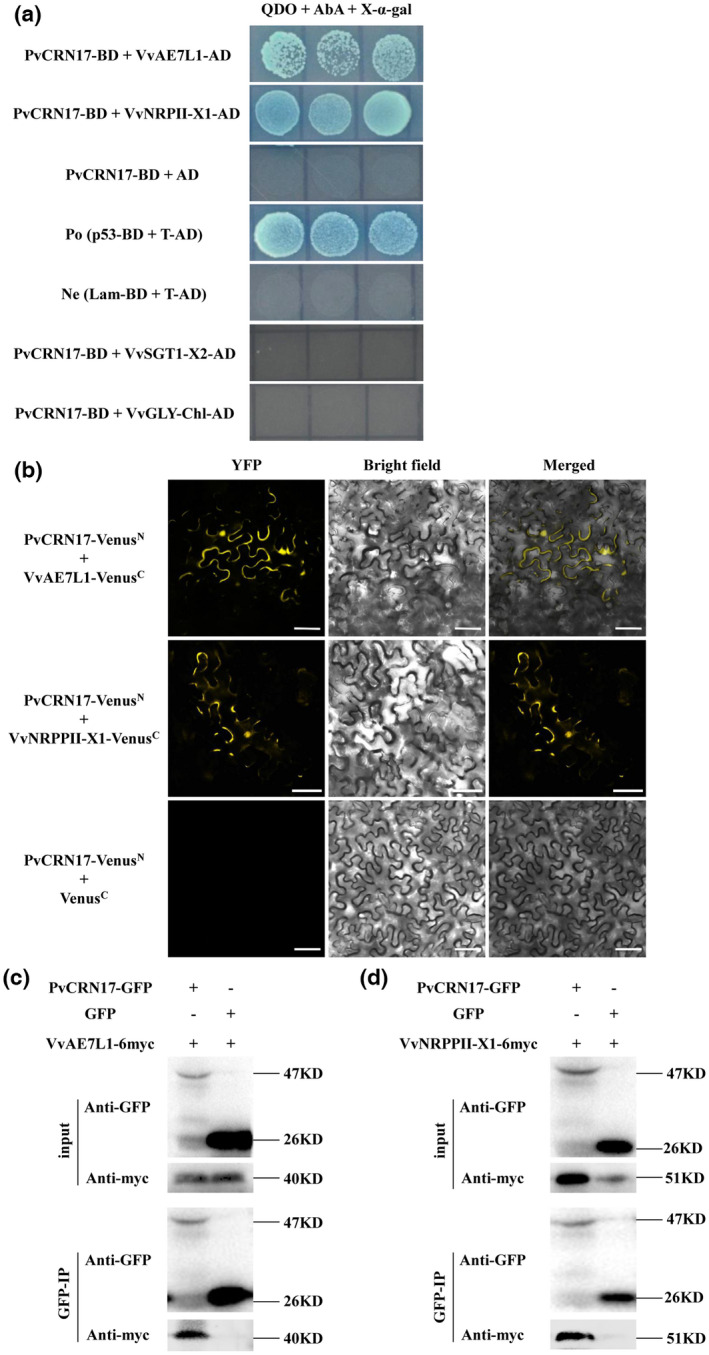
PvCRN17 interacts with VvAE7L1 and VvNRPPII‐X1 in vivo. (a) The interactions between PvCRN17 and VvAE7L1 as well as VvNRPPII‐X1 were validated by yeast two‐hybrid assay. (b) Bimolecular fluorescence complementation assay showed PvCRN17 interacted with VvAE7L1 and VvNRPPII‐X1. (c, d) Verification of the interaction between PvCRN17 and VvAE7L1 or VvNRPPII‐X1 by co‐immunoprecipitation
Sequence analysis showed that the sequences of the Vitis AE7L1 orthologous protein from V. vinifera ‘Thompson Seedless’ and ‘Pinot Noir’, V. piasezkii ‘Liuba‐8’, and V. riparia are identical (Figure S2 and Table S2). Therefore, most of the grapevine AE7‐Like 1 proteins (VAE7L1) are targets of PvCRN17. In addition, VvAE7L1 is the ortholog of AtAEL1 in A. thaliana and NbAE7L1 in N. benthamiana. (Figure S3). The interaction between PvCRN17 and AtAEL1 (UniProt accession number A8MR89) or NbAE7L1 (Sol Genomics Network accession number Niben101Scf02915g01001.1) was validated by Y2H, BiFC and Co‐IP assays. (Figure S4).
2.3. Both VAE7L1 and VAE7 are components of the CIA mechanism in grapevine
In plants, protein AE7‐Like 1 genes are paralogs of AE7 (ASYMMETRIC LEAVES1/2 ENHANCER 7) genes. In the genome of V. vinifera ‘PN40024’, the protein AE7‐Like 1 gene (LOC100259830) is the only paralog of AE7 (LOC100263182), and their coded proteins share 64.0% similarity. However, there are few reports describing the function of plant AE7 and AE7‐Like 1. Plant AE7 proteins contain the conserved DUF59 domain (NCBI accession TIGR02945), and AtAE7 is involved in the maturation of Fe‐S proteins as it interacts with the key components of the CIA pathway, AtCIA1 and AtMET18 (Luo et al., 2012). Here, conserved domain analysis indicates that VAE7L1, VvAE7, NbAE7L1, AtAEL1 and AtAE7 all belong to the FeS_assembly_P Superfamily (NCBI accession cl00941) (Figure S5), suggesting that conserved characteristics are shared by these proteins. According to the genome information of V. vinifera ‘PN40024’ and V. riparia ‘Gloire de Montpellier’, the CIA mechanisms could be conserved between grapevine and Arabidopsis, as every orthologous gene in grapevine of a single copy consensus to each gene in the CIA pathway in Arabidopsis was found (Table 1). Despite lacking genetic analysis, the interactions between VAE7L1/VvAE7 and VvCIA1 in vivo were verified in this study (Figure 4a,b). It demonstrates that VAE7L1 and VAE7 are working partners of VCIA1. A previous study showed that AtAE7 interacts with AtMET18 by directly binding to the C‐terminal region of AtMET18 (Luo et al., 2012). Here, VvAE7 also interacted with the C‐terminal region of VvMET18 (VvMET18‐C) and AtMET18‐C, respectively. AtAEL1 was found to interact with AtMET18. However, VAE7L1 failed to interact with VvMET18‐C and VvMET18‐N (Figure S6). The interaction between VAE7L1 and VvMET18 remains a further verification under the premise of the access to the complete coding fragment of VvMET18. Above all, plant AE7 and AE7‐Like 1 all correlate with the CIA mechanism. A further study showed that in grapevine and Arabidopsis, AE7‐Like 1 and AE7 interacted with each other (Figure 4c,d), indicating that AE7‐Like 1 and AE7 could make up a dimer. Plant AE7‐Like 1 and AE7 probably synergistically assist the delivery of Fe‐S clusters to precursor Fe‐S proteins.
TABLE 1.
Several conserved genes involved in the cytosolic iron‐sulphur cluster assembly (CIA) pathway in two Vitis species
| Arabidopsis thaliana | Vitis vinifera ‘PN40024’ | V. riparia ‘Gloire de Montpellier’ | |||
|---|---|---|---|---|---|
| Gene | Locus tag | Ortholog | Locus tag | Ortholog | Locus tag |
| NAR1 | At4g16440 | VIT_00s0508g00020 | LOC100267779 | Protein NAR1 | LOC117924258 |
| CIA1 | At2g26060 | VIT_08s0007g03360 | LOC100256192 | Protein CIA1 | LOC117920801 |
| AE7 | At1g68310 | VIT_08s0007g01580 | LOC100263182 | Protein AE7 | LOC117920429 |
| MET18 | At5g48120 | VIT_09s0002g03490 | LOC100260012 | MET18 | LOC117922349 |
| NBP35 | At5g50960 | VIT_14s0171g00460 | LOC100259370 | NBP35 | LOC117930919 |
FIGURE 4.
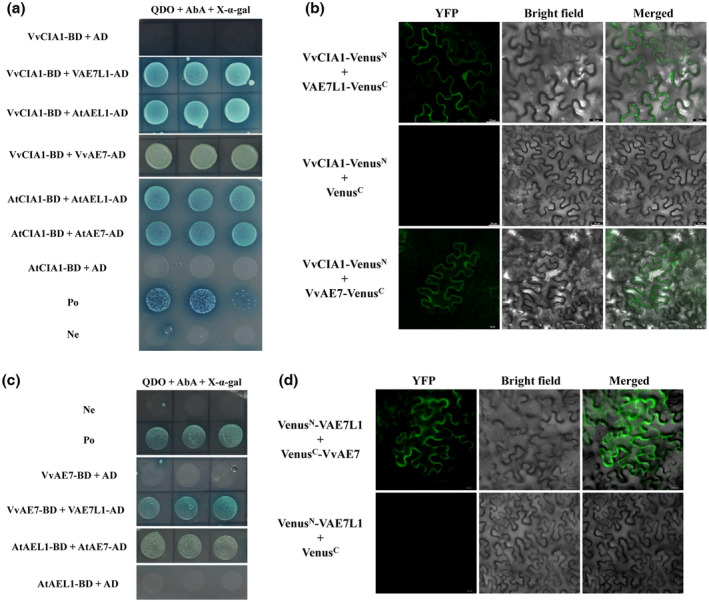
Both VAE7L1 and VvAE7 are the components of the plant cytosolic iron‐sulphur cluster assembly (CIA) pathway. (a) Yeast two‐hybrid (Y2H) assay showed that both VAE7L1 and VvAE7 interacted with VvCIA1. (b) Bimolecular fluorescence complementation (BiFC) assay showed that both VAE7L1 and VvAE7 interacted with VvCIA1. (c, d) Y2H assay and BiFC assay suggest that VAE7L1 interacted with VvAE7
2.4. VAE7L1 affects plant disease resistance
The function of plant AE7‐Like 1 in plant–pathogen interactions is rarely documented. Therefore, the expression of AE7L1 in two grapevine genotypes challenged with P. viticola was measured. The expression level of VvAE7L1 in Pinot Noir leaves inoculated with P. viticola was similar to that of the control (mock‐treated leaves) at 0–72 h postinoculation (hpi) (Figure 5a). At 96 hpi, the relative expression level of VvAE7L1 in the inoculated Pinot Noir leaves was significantly higher than that of the control. At 120 hpi, no significant difference was found. However, the expression profile of VpAE7L1 in the highly resistant V. piasezkii ‘Liuba‐8’ leaves challenged with P. viticola was different. The expression level of VpAE7L1 at 48, 72, 96 and 120 hpi was significantly higher than that of the control at the same time points, respectively (Figure 5b). Therefore, Vitis AE7‐Like 1 genes were induced during the interaction between grapevine and P. viticola. Continuous high expression of AE7‐Like 1 during the development of grapevine downy mildew in the resistant genotype indicates that VAE7L1 is involved in the defence response of grapevine to P. viticola.
FIGURE 5.
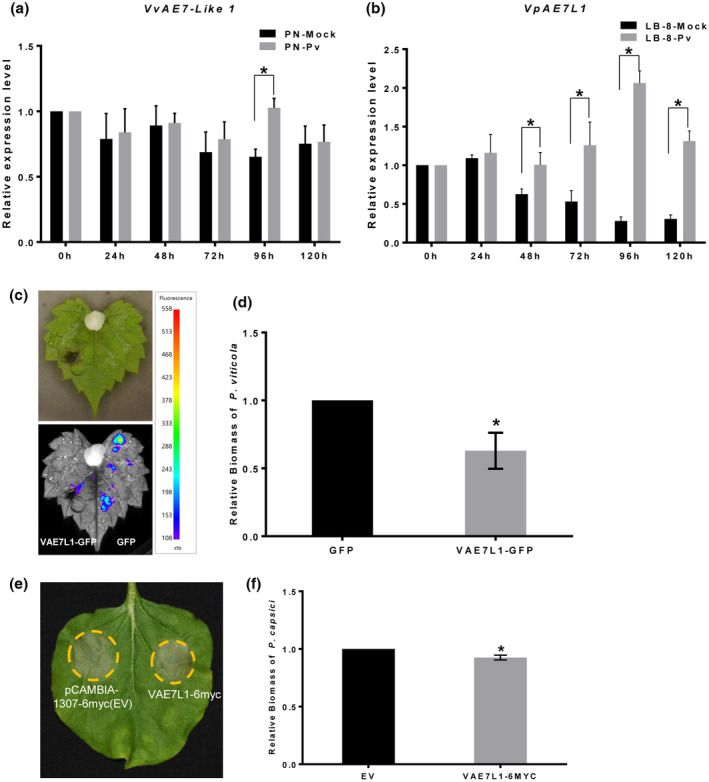
VAE7L1 is beneficial to the disease resistance of plants to pathogens. (a, b) VvAE7L1 in Vitis vinifera ‘Pinot Noir’ and VrAE7L1 in Vitis piasezkii ‘Liuba‐8’ were up‐regulated at some times during the development of grapevine downy mildew. Error bars represent the SEM. Asterisks indicate significant differences from the control groups (multiple t test; *p < 0.05). (c, d) Transient overexpression of VAE7L1 enhanced the resistance of V. riparia leaves to Plasmopara viticola. (e, f) Transient expression of VAE7L1 enhanced the resistance of Nicotiana benthamiana to Phytophthora capsici. The bars represent the SD. Asterisks indicate significant differences from the control groups (paired t test; *p < 0.05)
The effect of VAE7L1 on the resistance of grapevine to P. viticola was examined by inoculation of P. viticola on in vitro V. riparia leaves transiently expressing VAE7L1‐GFP (GFP served as the control) (Figure 5c). The average relative biomass of P. viticola in the leaves expressing VAE7L1‐GFP was smaller than that of the control (Figure 5d), suggesting that VAE7L1 enhanced the resistance of V. riparia leaves to P. viticola.
To further validate the positive role of VAE7L1 in regulating plant defence responses, N. benthamiana leaves transiently expressing VAE7L1‐6myc and the control (empty vector, EV) were inoculated with P. capsici. For leaves transiently expressing VAE7L1‐6myc, the disease symptoms were reduced and the average relative biomass of P. capsici in the lesions was smaller compared to that of the control (Figure 5e,f). NbAE7L1‐6myc was also transiently expressed in N. benthamiana leaves to evaluating its effect on the host resistance to P. capsici. However, there was no difference between the quantity of P. capsici in the leaves expressing NbAE7L1‐6myc and that of the control. Intriguingly, transient expression of AtAEL1‐GFP in N. benthamiana leaves significantly reduced the disease severity in the host plant (Figure S7). Thus, some FeS_assembly_P family proteins such as VAE7L1 and AtAEL1 contribute to the disease resistance of plants.
2.5. Some grapevine Fe‐S proteins positively regulate plant defence
The inference that VAE7L1 acts as one component in the CIA pathway suggests that VAE7L1 plays an indirect role in the defence system of grapevine, by fine‐tuning the delivery of Fe‐S clusters and the maturation of Fe‐S proteins. Then the expression of the CIA‐related genes, VvCIA1 and VvMET18, and four predicted Fe‐S protein coding genes, VvABCE2 (VIT_02s0087g00770), VvACO‐like‐12S (VIT_12s0059g02150, VvACOL‐12S), VvACO‐like‐19S (VIT_19s0090g00460, VvACOL‐19S) and VvDEMETER‐Like‐08S (VIT_08s0007g03920, VvDML‐08S), in Pinot Noir leaves challenged with P. viticola, were monitored. There was no difference between the expression of VvCIA1 in the Pinot Noir leaves inoculated with P. viticola and that of the control. However, the expression level of VvMET18 in the inoculated leaves at 96 and 120 hpi was increased compared to that of the control. Therefore, VvMET18 might correlate with the interaction between grapevine and P. viticola. For VvABCE2, its expression under the development of grapevine downy mildew was up‐regulated from 24 to 72 hpi. At the late stage of the disease (96 to 120 hpi) its expression in the infected leaves was similar to that of the control. The expression of VvACOL‐12S was up‐regulated from 24 hpi to 120 hpi, while the expression of VvACOL‐19S in the infected leaves showed higher levels at 24 hpi, 48hpi and 96 hpi. However, the expression of VvDML‐08S appeared uniform between the infected leaves and the control, stayping at low levels throughout the course of the disease (Figure S8).
VvABCE2, VvACOL‐12S and VvACOL‐19S, as well as VvFd2‐Like (VvFd2L, LOC100249080), were then cloned for further study. These genes were transiently expressed in N. benthamiana leaves, along with GFP expressed as the control (Figure S9). N. benthamiana leaves expressing these genes were inoculated with P. capsici zoospores for disease severity comparison. The lesions on N. benthamiana leaves where VvABCE2‐GFP was transiently expressed were significantly smaller than those of the control (Figure 6a). However, despite VvACOL‐12S being up‐regulated continuously in response to P. viticola, VvACOL‐12S‐GFP did not significantly enhance the resistance of N. benthamiana leaves to P. capsici (Figure 6b). Intriguingly, with the expression of VvACOL‐19S‐GFP, N. benthamiana leaves exhibited smaller lesions compared to the leaves transiently expressing GFP (Figure 6c). In summary, VABCE2 and VACOL‐19S could positively regulate plant defence reactions towards some oomycete pathogens. Consistent with the previous study on Arabidopsis (Wang et al., 2018), VvFd2L enhanced the resistance of N. benthamiana leaves to P. capsici (Figure 6d).
FIGURE 6.
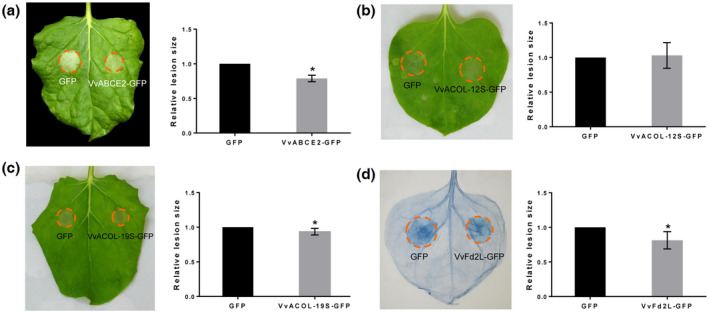
Disease resistance evaluation of Nicotiana benthamiana leaves transiently expressing Fe‐S coding genes VvABCE2 (a), VvACOL‐12S (b), VvACOL‐19S (c), and VvFd2L (d). Left panel: representative inoculated N. benthamiana leaves expressing the indicated constructs. Right panel: relative lesion size of the indicated infected sites. The error bars represent the SD. Asterisks indicate significant differences from the control groups (paired t‐test; *p < 0.05)
2.6. PvCRN17 competes with VvCIA1 to bind with VAE7L1 and VvAE7
As PvCRN17 interacts with VAE7L1, which interacts with VCIA1, there could be a competition between PvCRN17 and VCIA1 for the interaction with VAE7L1. Therefore, a competitive binding assay was conducted to verify the hypothesis. Firstly, PvCRN17 was shown not to interact with VvCIA1 directly in vivo by Y2H and BiFC assay (Figure 7a,b). Meanwhile, the interaction between VvCIA1 and VAE7L1 was confirmed by Co‐IP assay (Figure 7c). Then a competitive Co‐IP assay was performed. The amount of VAE7L1‐6myc co‐precipitated with VvCIA1‐GFP under the coexpression of PvCRN17‐mCherry was notably less than that of the control, in which mCherry served as the possible competitor (Figure 7d). This suggests that PvCRN17 could compete with VCIA1 to bind with VAE7L1 in the plant cell.
FIGURE 7.
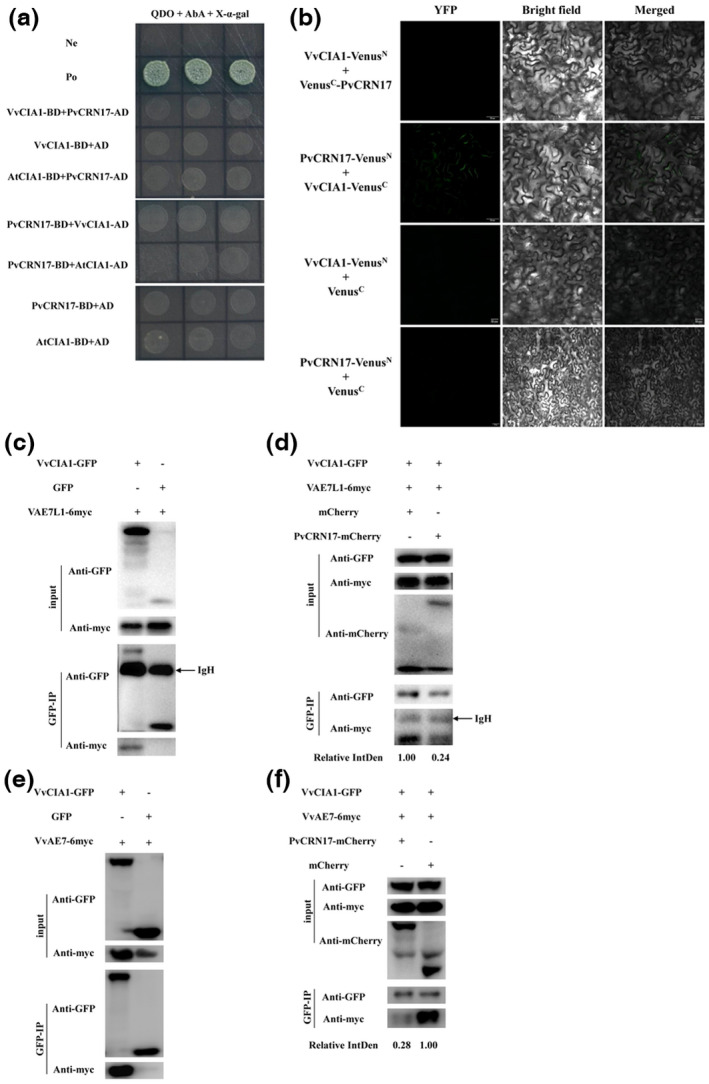
PvCRN17 competes with VvCIA1 to bind with VAE7L1 and VvAE7. (a, b) PvCRN17 could not interact with VvCIA1 directly in vivo. (c, d) Competitive co‐immunoprecipitation (Co‐IP) assay showed that PvCRN17 competed with VvCIA1 to bind with VAE7L1. (e) Validation of the interaction between VvAE7 and VvCIA1 by Co‐IP assay. (f) Competitive Co‐IP assay showed that PvCRN17 competed with VvCIA1 to bind with VvAE7. Relative IntDen indicates relative integrated density of the signal of VAE7L1‐6myc or VvAE7‐6myc precipitated with VvCIA1
Considering that VAE7 is the homologous protein of VAE7L1, the interaction between PvCRN17 and VvAE7 was also examined by Y2H and BiFC experiments. As expected, PvCRN17 interacted with VvAE7 and AtAE7 (Figure S10). Then, another competitive Co‐IP assay was conducted to verify the interfere by PvCRN17 in the interaction between VvCIA1 and VvAE7. The amount of VvAE7‐6myc co‐precipitated with VvCIA1‐GFP was reduced when PvCRN17 was introduced into the interaction system (Figure 7e,f). These findings suggest that PvCRN17 could compete with VCIA1 to bind with VAE7 as well as VAE7L1.
2.7. VAE7L1 could be trapped in the plasma membrane by PvCRN17
AtCIA1, AtAE7 and AtMET18 all reside in the cytosol and nucleus of the A. thaliana cell, making up a protein complex. The structures and functions of these proteins are considered to be conserved across the plant kingdom (Balk & Pilon, 2011; Luo et al., 2012). In this study, VAE7L1 was located to the cytosol and nucleus of the plant cell (Figure S11). However, colocalization analysis showed that VAE7L1 and PvCRN17 were mainly colocalized to the plasma membrane of the N. benthamiana cells referring to the colocalization ratio (Figure S11). With VAE7L1‐YFPN, VvAE7‐YFPC and PvCRN17‐mCherry coexpressed in N. benthamiana leaves, VAE7L1‐VvAE7 dimer and PvCRN17 also colocalized to the plasma membrane and the nucleus of the plant cell (Figure S11). Thus, VAE7L1 and VAE7 located in the cytoplasm could be tied to the plasma membrane by PvCRN17 when PvCRN17 is delivered into the grapevine cell. Interestingly, PvCRN17‐tmh, which turns to occupy the cytoplasm of the host cell, also interacted with VAE7L1 (Figure S12). Therefore, the transmembrane helix region has little effect on the interaction between PvCRN17 and VAE7L1.
3. DISCUSSION
CRN family genes comprise a class of ancient genes widely distributed in microorganisms. Like RXLR effectors, CRN effectors are intracellular‐targeted secretory proteins; for most of them, their functions remain to be uncovered (Amaro et al., 2017). In this study, a suspected virulent CRN‐like gene, PvCRN17, from P. viticola isolate YL, was confirmed to impair the resistance of host plants to pathogens. PvCRN17 could be destructive to the plant cell as PvCRN17‐transgenic Arabidopsis plants showed severe growth restriction. With the transcription of PvCRN17 in P. viticola detected during the development of the grapevine downy mildew (Xiang et al., 2021), these findings indicate that PvCRN17 is one strategic effector of P. viticola for exerting pathogenicity. It would be interesting to know how PvCRN17 works and which biological process is modulated by it in the host. Thus, the grapevine target proteins of PvCRN17 were explored in this study. VvAE7L1 and VvNRPPII‐X1 were both validated to interact with PvCRN17. Obviously, there are quite a few biological processes for PvCRN17 to affect. VvAE7L1 is an ortholog of AtAEL1 of A. thaliana, and the coded proteins of Vitis AE7‐like 1 gene alleles in four different grapevine genotypes are identical. Moreover, VAE7L1, AtAEL1 and NbAE7L1, as well as their homologous proteins VvAE7 and AtAE7, are FeS_assembly_P Superfamily proteins, of which the roles in plant defence responses remain to be investigated. Intriguingly, these proteins were all verified to interact with PvCRN17 in vivo. This suggests that PvCRN17 could influence a conserved biological process with pivotal roles in the host plant. Transient expression of VAE7L1 in the leaves of in vitro V. riparia plants increased the host resistance to P. viticola. Therefore, VAE7L1 could play an important role in the grapevine immunity system. In contrast to NbAE7L1, VAE7L1 transiently expressed in N. benthamiana markedly increased the host resistance to P. capsici, which supports the importance of VAE7L1 in regulating plant defence. During the development of grapevine downy mildew, the expression of AE7L1 in the leaves of the highly resistant V. piasezkii ‘Liuba‐8’ was up‐regulated earlier and remained at a higher level until 120 hpi, in contrast to the almost constant expression of AE7L1 in the leaves of the susceptible Pinot Noir. T provides clues to reveal the molecular mechanism under the differentiation of the resistance of Vitis genus to P. viticola.
Before this study, there were few reports concerning the contribution of plant AE7‐Like 1 gene to plant disease resistance. The signalling pathway VAE7L1 uses to enhance the resistance of the grapevine to P. viticola remains to be resolved. VAE7L1 is homologous to VvAE7 and AtAE7, which are evidently indispensable members of the CIA mechanisms in Vitis species and Arabidopsis, according to the published data (Bastow et al., 2017; Luo et al., 2012). Then the possibility comes up that VAE7L1 would interact with VCIA1 as well. Subsequently, these conjectures were verified, demonstrating that both VAE7L1 and VAE7 are involved in the CIA mechanism in grapevine. Besides, VAE7L1 was found to interact with VAE7 in vivo. Therefore, VAE7L1 and VAE7 are both components of the grapevine CIA mechanism, instead of acting as alternative members with redundant functions. In other words, VCIA1, VAE7L1, VAE7 and VMET18 together make up a large complex via physical interaction, facilitating the transfer of Fe‐S clusters to apoproteins to generate mature Fe‐S proteins. The complex could be conserved between grapevine and Arabidopsis as revealed by this study.
In N. benthamiana and A. thaliana, some members involved in the three Fe‐S cluster assembly pathways, along with several Fe‐S proteins, exert positive effects on plant resistance to nonhost and host pathogens. For example, AtNFS1(NITROGEN FIXATION S‐LIKE 1), which is a sulphur donor, together with its interactor AtFH (FRATAXIN), which is a predicted iron donor, contribute to the resistance of A. thaliana to the pathogen Pseudomonas syringae pv. tomato DC3000 as well as to two nonhost bacteria (Fonseca et al., 2020). The Fe‐S protein AtMFDX1 (Mitochondrial Ferredoxin‐1) also positively regulates plant defence to pathogens by accumulation of defence‐related metabolites (Fonseca et al., 2021). The chloroplast stromule‐localized Fe‐S protein in A. thaliana, Fd2 (Ferredoxin 2), was shown to be beneficial to plant resistance to a biotrophic pathogen as well as a hemibiotrophic pathogen (Wang et al., 2018). Besides, one N. benthamiana aconitase was found to be required for the cell death induction triggered by Bax and the interaction between Pto and the effector AvrPto (Moeder et al., 2007). In this study, the expression of three different Fe‐S proteins' coding genes, namely VvABCE2, VvACOL‐12S, VvACOL‐19S, as well as the CIA pathway‐related VvMET18 in Pinot Noir leaves challenged with P. viticola was up‐regulated. In addition, when transiently expressed, both VvABCE2 and VvACOL‐19S significantly enhanced the resistance of N. benthamiana leaves to P. capsici. The homologous protein of AtFd2 in grapevine, VvFd2L, when transiently expressed in N. benthamiana, also restricted the expansion of P. capsici in the leaves. Therefore, it is possible that the Fe‐S proteins studied here are conducive to the defence system of grapevine in response to P. viticola. However, the mechanisms by which these Fe‐S proteins regulate plant defence responses remain to be worked out in the future. There are further plant Fe‐S proteins remaining to be investigated for their roles in plant–pathogen interactions. Indirect evidence suggests that some Fe‐S proteins, such as DEMETER‐like proteins (DMLs) and abscisic aldehyde oxidases (AAOs), are favourable to plant disease resistance, as they could function upstream of the documented defence signalling pathways (Allègre et al., 2007; Le et al., 2014; Ton et al., 2009; Yu et al., 2013). Now, PvCRN17 could interact with VAE7L1 and VAE7, competing with VCIA1, and PvCRN17 was shown to bind firmly to the plant plasma membrane via its transmembrane domain. It is likely that when translocated into the grapevine cell, PvCRN17 could bind with VAE7L1 and VAE7, and anchor them to the plasma membrane, disrupting the interaction between VCIA1 and the VAE7L1‐VAE7 dimer. That would break up the Fe‐S clusters delivery complex formed by VNAR1, VCIA1, VAE7L1, VAE7 and VMET18, followed the failure of the integration of Fe‐S clusters into the apoproteins, causing function insufficiency or inactivation of the downstream Fe‐S proteins (Duan et al., 2015; Luo et al., 2012; Mõttus et al., 2021; Wang et al., 2016). As a result, PvCRN17 successfully suppressed the defence responses mediated by some Fe‐S proteins of grapevine (Figure 8). The model for the mechanism by which PvCRN17 inhibits the immune responses of grapevine deduced may provide a foundation for developing other effective strategies to control grapevine downy mildew. A particularly promising approach to improve the resistance of grapevine coudl be to look for or create novel alleles of Vitis AE7‐Like 1 or AE7 that encode proteins wh function in the CIA pathway but do not interact with virulent effectors from P. viticola.
FIGURE 8.
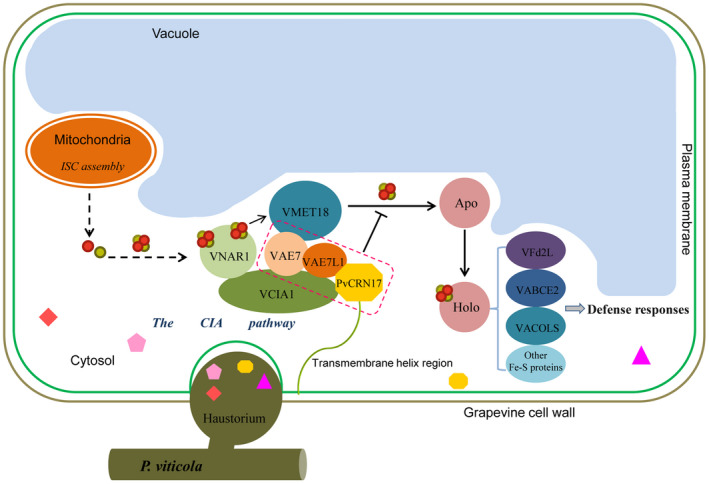
An proposed model for the mechanism by which PvCRN17 could interfere with the defence responses of the host grapevine. The small polygonal symbols represent the effectors secreted by Plasmopara viticola. The red and gold circles represent the elements Fe and S, respectively. Apo, apoproteins that are not conjugated with Fe‐S clusters; Holo, mature Fe‐S proteins that contain Fe‐S clusters. The ‘T' symbol represents inhibiting effect
4. EXPERIMENTAL PROCEDURES
4.1. Plant materials, microbes and culture conditions
Grapevine V. vinifera ‘Pinot Noir’, V. vinifera ‘Thompson Seedless’, V. riparia and V. piasezkii ‘Liuba‐8’ were grown in the Grape Germplasm Resources Repository at Northwest A & F University, Yangling, China. In vitro grown plantlets of V. riparia were propagated in half‐strength Murashige and Skoog medium (MS), 15 g/L sucrose, 0.15 mg/L indol‐3‐butyric acid (IBA), and 3 g/L phytagel. The in vitro plants in jars were grown under 24 ± 2°C in a tissue culture laboratory, and the photoperiod was 16 h illumination: 8 h dark. N. benthamiana plants and Arabidopsis plants were grown at 22 ± 2°C under a 16 h photoperiod and 75% relative humidity in a phytotron (Xiang et al., 2021).
Escherichia coli Top10 used for cloning was selectively cultured in solid or liquid Luria‐Bertani (LB) medium containing kanamycin (50 mg/L) at 37°C according to the donor plasmid used in this study. Agrobacterium tumefaciens GV3101 transformed with the indicated constructs were cultured at 28°C in solid or liquid LB medium supplemented with kanamycin (50 mg/L), gentamycin (30 mg/L) and rifampicin (50 mg/L). Yeast strain Y2H‐Gold used for yeast two‐hybrid assay was cultured at 30°C. The yeast cells containing the mixed cDNA library of Pinot Noir and V. piasezkii ‘Liuba‐8’ leaves challenged with P. viticola constructed previously were stored at −80°C.
P. viticola used for inoculation was originally collected from the leaves of Thompson Seedless with grapevine downy mildew in Yangling. The population of P. viticola was preserved on detached Thompson Seedless leaves by subculture with the sporangia (Mestre et al., 2012). P. capsici was maintained by inoculating the fresh mycelia on solid V8 juice medium (2.5% V8, 1.5% agar) at 25°C in the dark every 7–10 days (Song et al., 2015).
4.2. Nucleic acid preparation, cDNA synthesis, vector construction and reverse transcriptio‐quantitative PCR
The genomic DNA of plants or microbes was isolated with the CTAB method (Xin & Chen, 2012). Total RNA was isolated using a E.Z.N.A. Plant RNA Kit (Omega Bio‐tek) following the recommended protocol in the manual. The first‐strand cDNA was synthesized using a Primescript II 1st Strand cDNA Synthesis Kit (Takara) with the total RNA (2 μg per sample). The single‐strand cDNA was diluted 50‐fold with nuclease‐free water for gene cloning and real‐time quantitative PCR (qPCR).
The DNA fragments for cloning as indicated were amplified with specific primers (Table S3) from the cDNA templates or the corresponding diluted recombinant plasmids, using a KOD‐Plus‐Neo DNA Polymerase Kit (TOYOBO). The PCR product was digested with appropriate restriction enzymes and introduced into the linearized vector by ligation with T4 DNA ligase (Takara). Otherwise, the PCR product was directly inserted into the linearized vector by homologous recombination using the 2 × Seamless Cloning Mix (Biomed). All recombinant plasmids were confirmed by sequencing before use.
Gene expression at the transcriptional level was determined by reverse transcription (RT)‐qPCR with gene‐specific primers (Table S3) using the TransStart Top Green qPCR SuperMix (Transgen). The reactions were performed in the RT‐qPCR instrument Bio‐Rad iQ5 referring to the manufacturer's instructions. For grapevine genes' expression analysis, the Vitis Actin gene was used as the internal control. For N. benthamiana genes, NbEF1‐α served as the internal control. Statistical analysis was accomplished with Microsoft Excel and GraphPad Prism 6.
4.3. Transient expression and stable transformation of exogenous genes in plants
Transient expression of the interested gene in N. benthamiana leaves or in vitro V. riparia leaves was conducted by Agrobacterium‐infiltration as previously described (Xiang et al., 2021; Zottini et al., 2008). Stable genetic transformation of pYJ‐PvCRN17‐mCherry in A. thaliana plants was conducted with the floral dip method (Zhang et al., 2006).
4.4. Fluorescence imaging
The expression of indicated proteins tagged with GFP or mCherry transiently expressed in plants were confirmed by fluorescence detection with laser scanning confocal microscopy or fluorescence microscopy. In addition, transient expression of PvCRN17‐GFP and VAE7L1‐GFP in in vitro V. riparia leaves were detected by the PlantView 100 plant in vivo imaging system (Biolight) referring to the manufacturer's instructions.
4.5. Pathogen inoculation and disease resistance evaluation
Inoculation of P. capsici on N. benthamiana leaves was performed as the previous study described (Song et al., 2015; Xiang et al., 2021). The disease resistance of the leaf region expressing the indicated gene was measured by comparing the lesion size or the biomass of the P. capsici in the lesions to that of the control (Llorente et al., 2010; Xiang et al., 2021). For inoculating P. viticola on detached in vitro grapevine leaves infiltrated with A. tumefaciens containing indicated constructs (48 h later), equal amounts of droplets (10 μl each) of P. viticola sporangial suspension (50,000 sporangia/ml) were inoculated onto the left and right sides of each leaf on the abaxial surface. The inoculated leaves were then placed in the water‐agar medium, incubated in a plant growth chamber at 24 ± 2°C, relative humidity 75%, with a photoperiod of 16 h. The symptoms were recorded after the sporangiophores emerged. Disease resistance comparison was conducted by calculating the relative biomass of P. viticola in the leaves as indicated.
For the inoculation of P. viticola on detached grapevine leaves collected from the field, the leaves were sterilized with 1% sodium hypochlorite and sprayed with the sporangial suspension (50,000 sporangia/ml). Then, the inoculated leaves were placed in a tray padded with wet filter paper and incubated in the plant growth chamber with the same culture conditions as mentioned above. The experiment was conducted with three biological repeats for each group. Different batches of inoculated grapevine leaves were collected at 0, 24, 48, 72, 96, and 120 hpi and all stored at −80°C before further use.
4.6. H2O2 detection
The accumulation of H2O2 in plant leaves was detected by 3,3′‐diaminobenzidine (DAB) staining (Thordal‐Christensen et al., 1997), and the relative staining intensity was measured with ImageJ software.
4.7. Yeast two‐hybrid assay
The coding region of PvCRN17 was inserted into pGBKT7 vector (BD) to make the construct pGBKT7‐PvCRN17, which was subsequently transformed into the yeast Y2H‐Gold with the lithium acetate transformation method. After confirming that PvCRN17 did not have autoactivation activity in Y2H‐Gold (data not shown), PvCRN17 was used as the bait to screen target proteins from the mixed cDNA library of Pinot Noir and V. piasezkii ‘Liuba‐8’ leaves challenged with P. viticola. Yeast mating assay was performed following the standard protocol of the Matchmaker Gold Yeast Two‐Hybrid System (Clontech). The coding region of each candidate target was cloned from Pinot Noir leaves and introduced to pGADT7 vector (AD) for verification by co‐transformation of the prey and the bait to Y2H‐Gold. Transformants were selectively cultured on SD/−Leu−Trp (double dropout, DDO) medium and viable colonies were transferred onto the SD/−Ade/−His/−Leu/−Trp + AbA + X‐α‐gal (quadruple dropout, QDO + AbA+ X‐α‐gal) medium for interaction verification.
4.8. BiFC assay
BiFC assay was performed following the previous studies (Gehl et al., 2009; Walter et al., 2004). The fluorescence of Venus or yellow fluorescent protein (YFP) protein in N. benthamiana cells was detected by confocal microscopy or fluorescence microscopy.
4.9. Co‐IP assay and competitive Co‐IP assay
For Co‐IP assay, protein pairs of interest were transiently coexpressed in N. benthamiana leaves, and then extracted with the plant total protein extraction buffer (50 mM Tris–HCl pH 7.4, 150 mM sodium chloride, 10% glycerol, 0.1% Triton X‐100, 0.1% NP‐40, 0.25% sodium deoxycholate, 2 mM EDTA pH 8.0, 2 mM dithiothreitol [DTT], 2 mM PMSF and Protease Inhibitor Cocktail [Roche]). The extracted protein in liquid phase was incubated with anti‐GFP monoclonal antibody by gentle shaking at 4°C for 4 h. Afterwards, the solution was mixed with Protein A + G Sepharose Beads (7sea), followed by gentle shaking at 4°C for about 5 h. Protein complexes bound to the beads were detected with appropriate antibodies by western blot after being rinsed four times with the Co‐IP wash buffer (50 mM Tris–HCl pH 7.4, 150 mM sodium chloride, 10% glycerol, 0.1% Triton X‐100, 0.1% NP‐40, 2 mM EDTA pH 8.0, 2 mM DTT, 2 mM PMSF).
Competitive Co‐IP assay was performed as the previous study described (Kang et al., 2017), similarly to the Co‐IP assay. Briefly, two interacting proteins and one hypothetical contender protein were transiently expressed together in the N. benthamiana leaves, then a normal Co‐IP assay conducted to measure the impact of the contender protein on the known interaction. The relative integrated density of the signal of VAE7L1‐6myc or VvAE7‐6myc precipitated with VvCIA1 was measured by ImageJ software.
Supporting information
Figure S1 PvCRN17‐transgenic Arabidopsis plants show growth arrest phenotypes. (a) Identification of the antibiotic‐resistant Arabidopsis plants transformed with PvCRN17‐mcherry by PCR using the genomic DNA as templates. (b) The phenotypes of T1 generation PvCRN17‐transgenic Arabidopsis plants growing under normal conditions
Figure S2 Sequence identity analysis of Vitis AE7‐Like 1 orthologous genes from four grapevine genotypes. (a) Multiple sequence alignment of Vitis AE7‐Like 1 orthologous genes from four grapevine genotypes. (b) Multiple sequence alignment of four Vitis AE7‐Like 1 orthologous proteins. Multiple sequence alignment was performed with CLC Sequence Viewer 8.0
Figure S3 VvAE7L1, AtAEL1 and NbAE7L1 are orthologous proteins and share high sequence similarities. Orthologous groups were predicted with Inparanoid8 (http://InParanoid.sbc.su.se). Multiple‐sequence alignment was performed by DNAMAN, the background of the amino acid residues that are identical in all proteins aligned is marked in blue and the background of the amino acid residues that share similar characters is marked in cyan
Figure S4 Yeast two‐hybrid, bimolecular fluorescence complementation and co‐immunoprecipitation assays showed that PvCRN17 interacted with AtAEL1 and NbAE7L1
Figure S5 Plant AE7L1 share the conserved domain FeS_assembly_P Superfamily. The protein sequences of VAE7L1, VvAE7, AtAEL1, AtAE7 and NbAE7L1 were submitted to the NCBI‐CDD program to search for conserved domains in these proteins. A highly conserved domain FeS_assembly_P Superfamily (NCBI Accession: cl00941) was shared by these proteins
Figure S6 Validation of the interaction between VAE7L1 and VvMET18 by yeast‐two hybrid assay
Figure S7 The effects on the disease resistance of Nicotiana benthamiana leaves of transient expression of NbAE7L1 and AtAEL1. (a) NbAE7L1 transiently expressed in N. benthamiana failed to enhance the host resistance to Phytophthora capsici. (b) Transient expression of AtAEL1 in N. benthamiana leaves enhanced the host resistance to P. capsici
Figure S8 Relative expression level of two genes in the cytosolic iron‐sulphur cluster assembly CIA pathway (a, b) and four genes encoding Fe‐S proteins (c‐f) in Pinot Noir leaves challenged with Plasmopara viticola
Figure S9 Detection of four grapevine Fe‐S proteins transiently expressed in Nicotiana benthamiana leaves by fluorescence imaging. The fluorescence of VvABCE2‐GFP, VvACOL‐12S‐GFP, VvACOL‐19S‐GFP, VvFd2L‐GFP, and GFP were detected by laser confocal microscope 48 h after agroinfiltration
Figure S10 PvCR17 interacted with VvAE7 and AtAE7 as revealed by yeast two‐hybrid assay (a) and bimolecular fluorescence complementation (b)
Figure S11 Subcellular localization of VAE7L1 in the plant cell and colocalization of PvCRN17 and VAE7L1‐VvAE7 dimer in the plant cell. Upper panel: subcellular localization of VAE7L1 in Nicotiana benthamiana cells. Free mCherry was coexpressed with VAE7L1‐GFP to mark the cytoplasmic and nuclear distribution of proteins. Middle panel: colocalization analysis of PvCRN17 and VAE7L1 in N. benthamiana cells. Lower panel: colocalization analysis of PvCRN17 and the VAE7L1‐VvAE7 dimer. Recombinant proteins VAE7L1‐YFPN and VvAE7‐YFPC were coexpressed with PvCRN17‐mcherry in N. benthamiana leaves to analyse their colocalization. The fluorescence was detected by laser confocal microscope 48 h after agroinfiltration
Figure S12 Coimmunoprecipitation assay suggests that PvCRN17‐tmh, which is mainly distributed in the cytoplasm of the plant cell, interacts with VAE7L1
Table S1 The information of candidate prey proteins of PvCRN17 screened by yeast two‐hybrid assay
Table S2 Sequence information of grapevine AE7‐Like 1 orthologs cloned in this study
Table S3 Primers used in this study
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (nos. 31672115 and 31872054). All the authors declare that there is no conflict of interest.
Xiang, G. , Fu, Q. , Li, G. , Liu, R. , Liu, G. & Yin, X. et al. (2022) The cytosolic iron–sulphur cluster assembly mechanism in grapevine is one target of a virulent Crinkler effector from Plasmopara viticola . Molecular Plant Pathology, 23, 1792–1806. Available from: 10.1111/mpp.13266
Gaoqing Xiang, Qingqing Fu and Guanggui Li contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Allègre, M. , Daire, X. , Héloir, M.C. , Trouvelot, S. , Mercier, L. , Adrian, M. et al. (2007) Stomatal deregulation in Plasmopara viticola‐infected grapevine leaves. New Phytologist, 173, 832–840. [DOI] [PubMed] [Google Scholar]
- Amaro, T.M. , Thilliez, G.J. , Motion, G.B. & Huitema, E. (2017) A perspective on CRN proteins in the genomics age: evolution, classification, delivery and function revisited. Frontiers in Plant Science, 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk, J. & Lobréaux, S. (2005) Biogenesis of iron‐sulfur proteins in plants. Trends in Plant Science, 10, 324–331. [DOI] [PubMed] [Google Scholar]
- Balk, J. & Pilon, M. (2011) Ancient and essential: the assembly of iron‐sulfur clusters in plants. Trends in Plant Science, 16, 218–226. [DOI] [PubMed] [Google Scholar]
- Balk, J. & Schaedler, T.A. (2014) Iron cofactor assembly in plants. Annual Review of Plant Biology, 65, 125–153. [DOI] [PubMed] [Google Scholar]
- Bastow, E.L. , Bych, K. , Crack, J.C. , Le Brun, N.E. & Balk, J. (2017) NBP35 interacts with DRE2 in the maturation of cytosolic iron‐sulphur proteins in Arabidopsis thaliana . The Plant Journal, 89, 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier, J. , Przybyla‐Toscano, J. , Roret, T. , Didierjean, C. & Rouhier, N. (2015) The roles of glutaredoxins ligating Fe‐S clusters: sensing, transfer or repair functions? Biochimica et Biophysica Acta, 1853, 1513–1527. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. , Horvath, D.M. & Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, C.G. , Wang, X. , Tang, K. , Zhang, H. , Mangrauthia, S.K. , Lei, M. et al. (2015) MET18 connects the cytosolic iron‐sulfur cluster assembly pathway to active DNA demethylation in Arabidopsis . PLoS Genetics, 11, e1005559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, J.P. , Lee, H.K. , Boschiero, C. , Griffiths, M. , Lee, S. , Zhao, P. et al. (2020) Iron‐sulfur cluster protein NITROGEN FIXATION S‐LIKE1 and its interactor FRATAXIN function in plant immunity. Plant Physiology, 184, 1532–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, J.P. , Oh, S. , Boschiero, C. , Watson, B. , Huhman, D. & Mysore, K.S. (2021) The Arabidopsis iron‐sulfur (Fe‐S) cluster gene MFDX1 plays a role in host and nonhost disease resistance by accumulation of defense‐related metabolites. International Journal of Molecular Sciences, 22, 7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl, C. , Waadt, R. , Kudla, J. , Mendel, R.R. & Hänsch, R. (2009) New GATEWAY vectors for high throughput analyses of protein‐protein interactions by bimolecular fluorescence complementation. Molecular Plant, 2, 1051–1058. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H. , Handsaker, R.E. , Cano, L.M. et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Johnson, D.C. , Dean, D.R. , Smith, A.D. & Johnson, M.K. (2005) Structure, function, and formation of biological iron‐sulfur clusters. Annual Review of Biochemistry, 74, 247–281. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. & Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annual Review of Phytopathology, 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Kang, E. , Zheng, M. , Zhang, Y. , Yuan, M. , Yalovsky, S. , Zhu, L. et al. (2017) The microtubule‐associated protein MAP18 affects ROP2 GTPase activity during root hair growth. Plant Physiology, 174, 202–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, T.N. , Schumann, U. , Smith, N.A. , Tiwari, S. , Au, P.C. , Zhu, Q.H. et al. (2014) DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis . Genome Biology, 15, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill, R. & Mühlenhoff, U. (2008) Maturation of iron‐sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annual Review of Biochemistry, 77, 669–700. [DOI] [PubMed] [Google Scholar]
- Lill, R. (2009) Function and biogenesis of iron‐sulphur proteins. Nature, 460, 831–838. [DOI] [PubMed] [Google Scholar]
- Llorente, B. , Bravo‐Almonacid, F. , Cvitanich, C. , Orlowska, E. , Torres, H.N. , Flawiá, M.M. et al. (2010) A quantitative real‐time PCR method for in planta monitoring of Phytophthora infestans growth. Letters in Applied Microbiology, 51, 603–610. [DOI] [PubMed] [Google Scholar]
- Luo, D. , Bernard, D.G. , Balk, J. , Hai, H. & Cui, X. (2012) The DUF59 family gene AE7 acts in the cytosolic iron‐sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis . The Plant Cell, 24, 4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre, P. , Piron, M.‐C. & Merdinoglu, D. (2012) Identification of effector genes from the phytopathogenic oomycete Plasmopara viticola through the analysis of gene expression in germinated zoospores. Fungal Biology, 116, 825–835. [DOI] [PubMed] [Google Scholar]
- Miller, R.N. , Costa Alves, G.S. & Van Sluys, M.A. (2017) Plant immunity: unravelling the complexity of plant responses to biotic stresses. Annals of Botany, 119, 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder, W. , Del Pozo, O. , Navarre, D.A. , Martin, G.B. & Klessig, D.F. (2007) Aconitase plays a role in regulating resistance to oxidative stress and cell death in Arabidopsis and Nicotiana benthamiana . Plant Molecular Biology, 63, 273–287. [DOI] [PubMed] [Google Scholar]
- Mõttus, J. , Maiste, S. , Eek, P. , Truve, E. & Sarmiento, C. (2021) Mutational analysis of Arabidopsis thaliana ABCE2 identifies important motifs for its RNA silencing suppressor function. Plant Biology, 23, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou, B. , Jones, J. & Ding, P. (2022) Plant immune networks. Trends in Plant Science, 27, 255–273. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T. & Kemmerling, B. (2009) Chapter 1 PAMP‐triggered basal immunity in plants. In: Van Loon, L.C. (Ed.) Advances in botanical research, 51, 1–38. [Google Scholar]
- Peng, Y. , van Wersch, R. & Zhang, Y. (2018) Convergent and divergent signaling in PAMP‐triggered immunity and effector‐triggered immunity. Molecular Plant‐Microbe Interactions, 31, 403–409. [DOI] [PubMed] [Google Scholar]
- Pruitt, R.N. , Locci, F. , Wanke, F. , Zhang, L. , Saile, S.C. , Joe, A. et al. (2021) The EDS1‐PAD4‐ADR1 node mediates Arabidopsis pattern‐triggered immunity. Nature, 598, 495–499. [DOI] [PubMed] [Google Scholar]
- Rouault, T.A. (2019) The indispensable role of mammalian iron sulfur proteins in function and regulation of multiple diverse metabolic pathways. Biometals, 32, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack, S. , Huitema, E. , Cano, L.M. , Bozkurt, T.O. , Oliva, R. , Van Damme, M. et al. (2009) Ten things to know about oomycete effectors. Molecular Plant Pathology, 10, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack, S. , van Damme, M. , Bozkurt, T.O. , Cano, L.M. , Smoker, M. , Thines, M. et al. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proceedings of the National Academy of Sciences of the United States of America, 107, 17421–17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, T. , Ma, Z. , Shen, D. , Li, Q. , Li, W. , Su, L. et al. (2015) An oomycete CRN effector reprograms expression of plant HSP genes by targeting their promoters. PLoS Pathogens, 11, e1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W. , Forderer, A. , Yu, D. & Chai, J. (2021) Structural biology of plant defence. New Phytologist, 229, 692–711. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. & Dong, X. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nature Reviews Immunology, 12, 89–100. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. , Wei, Y. & Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. The Plant Journal, 11, 1187–1194. [Google Scholar]
- Tian, H. , Wu, Z. , Chen, S. , Ao, K. , Huang, W. , Yaghmaiean, H. et al. (2021) Activation of TIR signalling boosts pattern‐triggered immunity. Nature, 598, 500–503. [DOI] [PubMed] [Google Scholar]
- Ton, J. , Flors, V. & Mauch‐Mani, B. (2009) The multifaceted role of ABA in disease resistance. Trends in Plant Science, 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Tong, W.H. & Rouault, T.A. (2007) Metabolic regulation of citrate and iron by aconitases: role of iron‐sulfur cluster biogenesis. Biometals, 20, 549–564. [DOI] [PubMed] [Google Scholar]
- Walter, M. , Chaban, C. , Schütze, K. , Batistic, O. , Weckermann, K. , Näke, C. et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal, 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Rui, L. , Yan, H. , Shi, H. , Zhao, W. , Lin, J.E. et al. (2018) The major leaf ferredoxin Fd2 regulates plant innate immunity in Arabidopsis . Molecular Plant Pathology, 19, 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Li, Q. , Yuan, W. , Cao, Z. , Qi, B. , Kumar, S. et al. (2016) The cytosolic Fe‐S cluster assembly component MET18 is required for the full enzymatic activity of ROS1 in active DNA demethylation. Scientific Reports, 6, 26443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, G. , Yin, X. , Niu, W. , Chen, T. , Liu, R. , Shang, B. et al. (2021) Characterization of CRN‐Like Genes From Plasmopara viticola: searching for the most virulent ones. Frontiers in Microbiology, 12, 632047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Z. & Chen, J. (2012) A high throughput DNA extraction method with high yield and quality. Plant Methods, 8, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, X. , Shang, B. , Dou, M. , Liu, R. , Chen, T. , Xiang, G. et al. (2019) The nuclear‐localized RxLR effector PvAvh74 from Plasmopara viticola induces cell death and immunity responses in Nicotiana benthamiana . Frontiers in Microbiology, 10, 1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, A. , Lepère, G. , Jay, F. , Wang, J. , Bapaume, L. , Wang, Y. et al. (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proceedings of the National Academy of Sciences of the United States of America, 110, 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M. , Ngou, B. , Ding, P. & Xin, X.F. (2021) PTI‐ETI crosstalk: an integrative view of plant immunity. Current Opinion in Plant Biology, 62, 102030. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Henriques, R. , Lin, S.S. , Niu, Q.W. & Chua, N.H. (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols, 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zottini, M. , Barizza, E. , Costa, A. , Formentin, E. , Ruberti, C. , Carimi, F. et al. (2008) Agroinfiltration of grapevine leaves for fast transient assays of gene expression and for long‐term production of stable transformed cells. Plant Cell Reports, 27, 845–853. [DOI] [PubMed] [Google Scholar]
- Łaźniewska, J. , Macioszek, V.K. & Kononowicz, A.K. (2012) Plant‐fungus interface: the role of surface structures in plant resistance and susceptibility to pathogenic fungi. Physiological and Molecular Plant Pathology, 78, 24–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 PvCRN17‐transgenic Arabidopsis plants show growth arrest phenotypes. (a) Identification of the antibiotic‐resistant Arabidopsis plants transformed with PvCRN17‐mcherry by PCR using the genomic DNA as templates. (b) The phenotypes of T1 generation PvCRN17‐transgenic Arabidopsis plants growing under normal conditions
Figure S2 Sequence identity analysis of Vitis AE7‐Like 1 orthologous genes from four grapevine genotypes. (a) Multiple sequence alignment of Vitis AE7‐Like 1 orthologous genes from four grapevine genotypes. (b) Multiple sequence alignment of four Vitis AE7‐Like 1 orthologous proteins. Multiple sequence alignment was performed with CLC Sequence Viewer 8.0
Figure S3 VvAE7L1, AtAEL1 and NbAE7L1 are orthologous proteins and share high sequence similarities. Orthologous groups were predicted with Inparanoid8 (http://InParanoid.sbc.su.se). Multiple‐sequence alignment was performed by DNAMAN, the background of the amino acid residues that are identical in all proteins aligned is marked in blue and the background of the amino acid residues that share similar characters is marked in cyan
Figure S4 Yeast two‐hybrid, bimolecular fluorescence complementation and co‐immunoprecipitation assays showed that PvCRN17 interacted with AtAEL1 and NbAE7L1
Figure S5 Plant AE7L1 share the conserved domain FeS_assembly_P Superfamily. The protein sequences of VAE7L1, VvAE7, AtAEL1, AtAE7 and NbAE7L1 were submitted to the NCBI‐CDD program to search for conserved domains in these proteins. A highly conserved domain FeS_assembly_P Superfamily (NCBI Accession: cl00941) was shared by these proteins
Figure S6 Validation of the interaction between VAE7L1 and VvMET18 by yeast‐two hybrid assay
Figure S7 The effects on the disease resistance of Nicotiana benthamiana leaves of transient expression of NbAE7L1 and AtAEL1. (a) NbAE7L1 transiently expressed in N. benthamiana failed to enhance the host resistance to Phytophthora capsici. (b) Transient expression of AtAEL1 in N. benthamiana leaves enhanced the host resistance to P. capsici
Figure S8 Relative expression level of two genes in the cytosolic iron‐sulphur cluster assembly CIA pathway (a, b) and four genes encoding Fe‐S proteins (c‐f) in Pinot Noir leaves challenged with Plasmopara viticola
Figure S9 Detection of four grapevine Fe‐S proteins transiently expressed in Nicotiana benthamiana leaves by fluorescence imaging. The fluorescence of VvABCE2‐GFP, VvACOL‐12S‐GFP, VvACOL‐19S‐GFP, VvFd2L‐GFP, and GFP were detected by laser confocal microscope 48 h after agroinfiltration
Figure S10 PvCR17 interacted with VvAE7 and AtAE7 as revealed by yeast two‐hybrid assay (a) and bimolecular fluorescence complementation (b)
Figure S11 Subcellular localization of VAE7L1 in the plant cell and colocalization of PvCRN17 and VAE7L1‐VvAE7 dimer in the plant cell. Upper panel: subcellular localization of VAE7L1 in Nicotiana benthamiana cells. Free mCherry was coexpressed with VAE7L1‐GFP to mark the cytoplasmic and nuclear distribution of proteins. Middle panel: colocalization analysis of PvCRN17 and VAE7L1 in N. benthamiana cells. Lower panel: colocalization analysis of PvCRN17 and the VAE7L1‐VvAE7 dimer. Recombinant proteins VAE7L1‐YFPN and VvAE7‐YFPC were coexpressed with PvCRN17‐mcherry in N. benthamiana leaves to analyse their colocalization. The fluorescence was detected by laser confocal microscope 48 h after agroinfiltration
Figure S12 Coimmunoprecipitation assay suggests that PvCRN17‐tmh, which is mainly distributed in the cytoplasm of the plant cell, interacts with VAE7L1
Table S1 The information of candidate prey proteins of PvCRN17 screened by yeast two‐hybrid assay
Table S2 Sequence information of grapevine AE7‐Like 1 orthologs cloned in this study
Table S3 Primers used in this study
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


