Abstract
Nonspecific lipid transfer proteins (LTPs) are small, cysteine‐rich proteins that play numerous functional roles in plant growth and development, including cutin wax formation, pollen tube adhesion, cell expansion, seed development, germination, and adaptation to changing environmental conditions. LTPs contain eight conserved cysteine residues and a hydrophobic cavity that provides a wide variety of lipid‐binding specificities. As members of the pathogenesis‐related protein 14 family (PR14), many LTPs inhibit fungal or bacterial growth, and act as positive regulators in plant disease resistance. Over the past decade, these essential immunity‐related roles of LTPs in plant immune processes have been documented in a growing body of literature. In this review, we summarize the roles of LTPs in plant–pathogen interactions, emphasizing the underlying molecular mechanisms in plant immune responses and specific LTP functions.
Keywords: lipid transfer protein, plant, plant disease, systemic acquired resistance, pathogen
This article summarizes the roles of lipid transfer proteins (LTPs) in plant–pathogen interactions, emphasizing the underlying molecular mechanisms in plant immune responses and specific LTP functions.
1. INTRODUCTION
Nonspecific lipid transfer proteins (LTPs) were first isolated from potato tubers and identified by Kader (1975). LTPs are ubiquitous small proteins (molecular weights ranging from 6.5 to 10 kDa) that exist in all terrestrial plants (Salminen et al., 2016). They have eight cysteine residue motifs that form four conserved disulphide bridges, which help stabilize the peptide tertiary structure, and many α‐helices with a central hydrophobic cavity capable of binding to a variety of lipids such as fatty acids, fatty acyl‐CoA, phospholipids, and prostaglandin B2 (Madni et al., 2020; Salminen et al., 2016). LTPs are encoded by large gene families and expressed abundantly in a variety of plant tissues. Almost all nonspecific LTPs carry an N‐terminal signal that localizes the protein at the subcellular level (Edstam et al., 2011; Missaoui et al., 2022). Traditionally, LTPs are classified according to molecular weight or sequence identity (Boutrot et al., 2008; Kalla et al., 1994). A more recent classification system by Edstam et al. (2011) uses five major types (LTP1, LTP2, LTPc, LTPd and LTPg) and five minor types (LTPe, LTPf, LTPh, LTPj and LTPk) based on the position of a conserved intron, the amino acid sequence identity, and posttranslational modifications. Expression of many nonspecific LTPs can be induced by multiple biotic and abiotic stresses, including disease, salinity, temperature and drought (Akhiyarova et al., 2021; Duo et al., 2021; Safi et al., 2015; Zhao et al., 2021). In the past two decades, several specific LTP functions have been identified, including defence against pathogens (Ben et al., 2021; Chen et al., 2021; McLaughlin et al., 2021; Schmitt et al., 2018), abiotic stress response (Dhar et al., 2020; Hairat et al., 2018; Zhao et al., 2020), pollen development (Andre et al., 2022, Tao et al., 2021), cutin wax formation (Debono et al., 2009; Kim et al., 2012; Lee et al., 2009; Liu et al., 2014), and seed development and germination (Wang et al., 2015). This evidence and the long evolutionary history of LTPs further corroborate their likely roles in plant stress adaptation and defence. For a detailed summary of LTP classification, three‐dimensional structures, and common functions in plant growth and development, see Salminen et al. (2016). In this review, we summarize current knowledge of LTPs in plant–pathogen interactions and discuss interesting future research directions.
2. ROLES OF LTPs IN DIRECT INHIBITION OF FUNGAL AND BACTERIAL GROWTH OR SPORE GERMINATION IN VIVO AND IN VITRO
Several LTPs have been found to directly inhibit the growth of pathogens in vitro. For example, recombinant Brassica rapa LTP2.1 displays direct antimicrobial activity against a wide range of plant pathogens (Schmitt et al., 2018), LTPs from Leonurus japonicus inhibited growth of filamentous fungi, bacteria and yeast in vitro (Yang et al., 2006), LTP4 from Triticum durum has a significant antibacterial effect against several gram‐positive and gram‐negative bacteria (Ben et al., 2021), recombinant Arabidopsis thaliana LTP4.4 exerts potent antifungal activity against Fusarium graminearum (McLaughlin et al., 2021), and LTPs from Gossypium hirsutum inhibit growth in a wide range of fungi (Chen et al., 2021).
As mentioned in the Introduction, LTPs contain four α‐helices that are stabilized by disulphide bonds formed by eight cysteine residues. LTPs are presumably able to bind lipids and other nonpolar substances due to these hydrophobic cavities. Several methods have been used to determine the three‐dimensional structures of various LTPs (Salminen et al., 2016). For example, in vitro lipid‐binding ability has been demonstrated in numerous LTPs, including those from B. rapa (LTP2.1), wheat and mung bean (Schmitt et al., 2018; Sun et al., 2008; Wang et al., 2004). Madni et al. (2020) analysed crystal structures of Solanum melongena LTPs to propose a lipid transport model in which the LTP N‐terminus binds lipids, resulting in the opening of the hydrophobic cavity. The lipids are then internalized into the hydrophobic cavity and expelled from the LTP C‐terminus (Figure 1). This model suggests that LTPs may help increase the porosity of fungal membranes by bleaching lipids.
FIGURE 1.
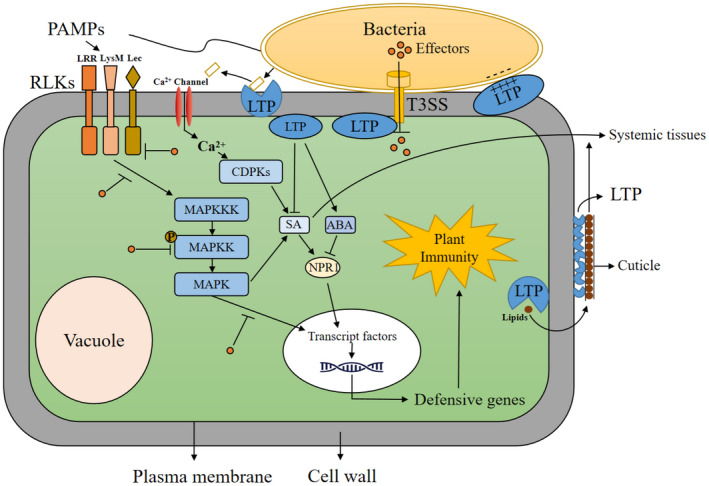
Lipid transfer protein (LTP) roles in plant and bacteria interactions. LTPs destroy bacterial cell membranes using lipid transfer properties or their highly conserved cationic residues. LTPs might directly bind to constituent proteins of the bacterial type III secretion system to inhibit bacteria and secrete effectors into host cells. LTPs may regulate expression of salicylic acid‐ and N‐hydroxy‐pipecolic acid‐dependent genes by regulating the homeostasis of abscisic acid and salicylic acid. LTPs also might regulate long‐distance transport of salicylic acid by participating in cuticle synthesis. Finally, LTPs contribute to pathogen resistance by maintaining integrity of adhesion between the cuticle and underlying cell wall. ABA, abscisic acid; CDPKs, calcium‐dependent protein kinases; LRR, leucine‐rich repeat; PAMPs, pathogen‐associated molecular patterns; RLKs, receptor‐like kinases; SA, salicylic acid; T3SS, type III secretion system.
Although the antibacterial properties of LTPs appear to be closely associated with lipid‐binding activity, in some cases the antifungal properties of some LTPs have not been strongly correlated with their lipid‐binding activity (Sun et al., 2008). Ace‐AMP1, an antifungal protein in onion seeds, shares some structural similarities with plant LTPs but does not bind lipids (Tassin et al., 1998). Rice LTP110 with defective lipid‐binding activity caused by site‐directed mutagenesis retains some antifungal activity (Ge et al., 2003). These findings suggest that LTPs have other antifungal mechanisms. The cationic residues of LTPs may be associated with growth inhibition of pathogens. For example, De Samblanx et al. (1997) hypothesized that positively charged patches generated by cationic residues on the LTP surface interact with specific negatively charged membrane domains of fungal pathogens, resulting in alteration and destabilization of the membrane structure (Figure 1). Subsequent in vitro studies have supported this hypothesis: highly conserved cationic residues of mung bean LTP1 contribute to its antimicrobial activity (Lin et al., 2005) and the antibacterial activity of purified wheat LTPs in vitro is correlated with several cationic residues (Sun et al., 2008).
In vitro studies indicate that phytopathogen growth is inhibited by numerous LTPs in a wide variety of plant species (Ben et al., 2021; Chen et al., 2021; McLaughlin et al., 2021). Overexpression of various LTPs promotes resistance to bacterial and fungal infections in plants (Ali et al., 2020; Sarowar et al., 2009; Schmitt et al., 2018). LTPs can also interact with other proteins in plant cells (Lim et al., 2016; Yu et al., 2013), possibly inhibiting pathogens by interacting with their target proteins. Microbial pathogens secrete a wide range of effectors capable of inactivating host proteins, or inhibiting expression of host pathogenesis‐related (PR) genes, via their secretion systems (Deng et al., 2017, Lasica et al., 2017). The type III secretion system, in particular, is involved in modulating numerous cellular processes, including cytoskeletal functions, membrane trafficking and cell death (Jing et al., 2021). Ali et al. (2020) demonstrated a resistance role of AtLTPg5 in resistance against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) in Arabidopsis, showing that ltpg5 plants are more susceptible to Pst DC3000, and that PATHOGENESIS‐RELATED GENE 1 (PR1) expression level decreases in Pst DC3000‐infected ltpg5 plants. Similarly, we observed reduced expression levels of many PR genes in P. syringae pv. maculicola‐infected ltp mutants (authors' unpublished data), suggesting the presence of PR gene expression inhibitors in mutants infected by phytopathogenic bacteria. A likely scenario is that plant LTPs directly bind to and inhibit constituent proteins of the bacterial secretion system. Loss of LTP function thus allows bacteria to secrete more effectors into host cells (Figure 1).
Many studies have demonstrated the ability of LTPs to inhibit microbial pathogen activity in vitro. Whether the same mechanisms underlie antimicrobial activity in vivo remains unclear. LTP overexpression has been repeatedly shown to enhance plant resistance to bacteria, fungi, viruses and arthropod pests; examples include LTPs in Arabidopsis (Ali et al., 2020), Capsicum annuum (Sarowar et al., 2009), Triticum aestivum (Zhu et al., 2012), and B. rapa (Schmitt et al., 2018). Loss of glycosylphosphatidylinositol‐anchored lipid transfer protein (LTPg)1, LTPg2, LTPg5, or LTPg6 function in Arabidopsis increases susceptibility to epidermal cell wall penetration by Blumeria graminis f. sp. hordei (Bgh) (Fahlberg et al., 2019). LTPg1 is located on papillae, and observations of cell wall reinforcement at attempted Bgh penetration sites suggest that LTPg1 transports wax monomers to enhance papillary cuticular thickness, thereby blocking Bgh infection. However, Bgh infection does not alter the composition of cuticular wax in ltpg1 mutants, suggesting that LTPg1 exerts a direct antimicrobial effect.
3. LTPs CONTRIBUTE TO PLANT RESISTANCE RESPONSES BY REGULATING CUTICULAR WAX ACCUMULATION
The plant cuticle, which consists of a lipophilic cutin polymer matrix and wax, is the primary barrier against pathogen invasion. Cuticular wax is composed of long‐chain fatty acids and their derivatives. The cuticle's mechanical strength and viscoelastic properties help prevent pathogen infection (Hoffmann‐Benning & Kende, 1994; Lee et al., 2009; Riederer & Schreiber, 2001). Plant LTPs are generally located on the cell walls, suggesting involvement in the transport of cutin monomers and wax (Pyee & Kolattukudy, 1995). In tobacco plants, cuticular wax accumulation is associated with increased LTP expression (Cameron et al., 2006). In Arabidopsis, LTPg1 plays a role in cuticular lipid accumulation (Debono et al., 2009; Lee et al., 2009). Loss of LTPg1 function alters cuticular lipid composition but not total wax or cutin monomer loads. Moreover, the cuticle of ltpg1 plants is disorganized and diffuse relative to the wild‐type, indicating that LTPg1 helps maintain the cuticular layer structure (Lee et al., 2009). LTPg2 and LTPg6 function similarly to LTPg1 in epicuticular wax accumulation (Kim et al., 2012; Lee et al., 2009). BraLTP1 is involved in epicuticular wax deposition in Brassica napus (Liu et al., 2014), and ltpg1 gene knockdown results in increased susceptibility to the fungal pathogen Alternaria brassicicola (Lee et al., 2009). The effects of these LTPs on wax accumulation in plants thus are evidently related to their ability to enhance pathogen resistance.
A recent study found that LTPs play a major structural role in maintaining the integrity of adhesion between the cuticle (mainly hydrophobic) and underlying cell wall (hydrophilic) (Jacq et al., 2017). Arabidopsis LTP2 is localized in the cell wall and has only a minor effect on cuticular composition. However, ltp2 plants have a disorganized ultrastructure and increased permeability at the cuticle–cell wall interface. In addition, LTP2 expression in Arabidopsis is induced by the necrotrophic fungus Botrytis cinerea and soil bacterium Agrobacterium tumefaciens (Chassot et al., 2007), further suggesting that LTP2 contributes to pathogen resistance by maintaining integrity between the cuticle and the cell wall (Figure 1).
In addition to its function as a physical barrier against microbial invasion, the cuticle plays important roles in defensive signalling and systemic acquired resistance (SAR) (Chassot et al., 2007; Xia et al., 2009, 2010, 2012). In A. thaliana, expression of a cell wall‐targeted fungal cutinase induces alteration of the cuticular structure and enhances resistance to B. cinerea. Resistance responses in cutinase‐expressing plants are associated with increased fungitoxic activity and expression of PR genes (LTPs, protein inhibitor gene families, peroxidases), but such responses are independent of salicylic acid (SA)‐, ethylene‐ and jasmonic acid (JA)‐mediated signal transduction pathways (Chassot et al., 2007). The cuticle also plays a critical role in SAR induction (Lim et al., 2020; Xia et al., 2009). Loss functions of cuticle synthesis‐related proteins, such as acyl carrier protein 4 and long‐chain acyl‐coA synthetase 2, result in compromised SAR (Xia et al., 2009). Another study found that an intact cuticle is necessary for active transport of SA, an essential defensive hormone (Lim et al., 2020). A cuticle‐defective mutant shows disruption of long‐distance transport of SA from SAR‐inducing tissues to systemic tissues, which reduces accumulation of pipecolic acid (an SAR inducer) in distal tissues. LTPs also appear to be involved in cuticle synthesis (Debono et al., 2009; Kim et al., 2012; Lee et al., 2009). Thus, it is reasonable to speculate that LTPs mediate broad‐spectrum defensive signalling by regulating lipid export from cuticle (Figure 1).
4. ROLE OF LTPs IN SYSTEMIC ACQUIRED RESISTANCE
SAR is an inducible defensive response of plants triggered by localized pathogen attack. It can induce immunity to subsequent pathogen infection in the whole plant within a few days (Schnake et al., 2020; Zhou et al., 2021). Some long‐distance signals synthesized at the infection site during SAR can be translocated to systemic tissues via phloem or volatilization to induce disease resistance (Chanda et al., 2011; Chaturvedi et al., 2012; Jung et al., 2009; Riedlmeier et al., 2017). Maldonado et al. (2002) reported that DEFECTIVE IN INDUCED RESISTANCE 1 (DIR1), an LTP, plays a role in the transmission of mobile SAR signals in Arabidopsis. DIR1 can bind lipids with high affinity (Lascombe et al., 2008), suggesting that mobile SAR signals contain lipids. Further studies confirmed that G3P or its lipid derivatives are mobile SAR signals (Chanda et al., 2011). DIR1 is translocated to systemic leaves via plasmodesmata in the presence of glycerol‐3‐phosphate (G3P) and, conversely, G3P translocation requires DIR1, indicating that SAR establishment depends on long‐distance transport of DIR1 in conjunction with G3P or its derivatives (Chanda et al., 2011). Another LTP, DIR‐like, is also translocated to systemic leaves via the phloem and in some cases DIR‐like substitutes for DIR1 (Champigny et al., 2013). In addition to G3P‐derived lipid signalling, DIR1 also participates in dehydroabietinal‐induced SAR (Figure 2), and a dir1 mutant was found to be defective in dehydroabietinal‐induced systemic immunity (Chaturvedi et al., 2012).
FIGURE 2.
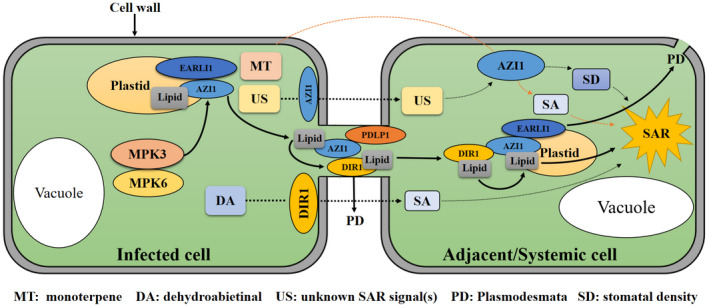
A schematic model describing the proposed functions for lipid transfer proteins (LTPs) in transporting lipid‐derived systemic acquired resistance (SAR) signals. During priming induction, MPK3/MPK6 promotes accumulation of AZI1 in plastids. AZI1/EARLI1 facilitates transport of SAR lipid signalling to the plasmodesmata (PD). Then, AZI1 interacts with plasmodesmata‐localizing protein 1 (PDLP1) and DIR1 in the plasmodesmata, transferring lipid signals to DIR1. The DIR1 and lipid signal complex is transferred to adjacent and systemic cells via the plasmodesmata. Lipid signalling then induces defence responses or transfer to AZI1 to further translocate SAR signals to other cells. Monoterpene (MT) induces a defence response via volatilized systemic tissues and neighbouring plants, where AZI1 mediates monoterpene‐induced defensive signal propagation. DIR1 is involved in dehydroabietinal (DA)‐induced SAR. AZI1 also might regulate systemic stomatal density (SD) via perception or transport of an unknown SAR signal (US).
Like DIR1, Azelaic Acid Induced 1 (AZI1) is an LTP that plays a similar role in the transfer of long‐distance SAR signalling. Loss of AZI1 function results in loss of systemic immunity triggered by pathogens or azelaic acid (AzA), a long‐distance SAR signal (Jung et al., 2009). AZI1 and DIR1 both mediate long‐distance transport of azelaic acid and G3P. DIR1 contains two Src homology 3 domains involved in promoting protein interactions (Gao et al., 2021; Lascombe et al., 2008), suggesting a possible AZI1–DIR1 interaction. The AZI1–DIR1 interaction has been confirmed by bimolecular fluorescence complementation and co‐immunoprecipitation (co‐IP) assays (Yu et al., 2013). AZI1 and DIR1 functions are essential for G3P accumulation; conversely, G3P regulates AZI1 and DIR1 transcription. These findings demonstrate that SAR is regulated by an intricate regulatory feedback loop among G3P, DIR1 and AZI1 (Yu et al., 2013).
Early Arabidopsis Aluminum Induced 1 (EARLI1) is an AZI1 homolog that can form complexes with AZI1 and DIR1, and is functionally involved in SAR (Cecchini et al., 2015). AZI1 and DIR1 are colocalized at perinuclear endoplasmic reticulum, plasmodesmata, and chloroplast/ endoplasmic reticulum contact sites. AZI1 also is located on plastids (e.g., intracellular endosymbiotic cyanobacteria, such as chloroplasts and chromoplasts), whereas DIR1 is not, indicating a distinction in their particular functions (Cecchini et al., 2021). Live cell imaging has revealed the presence of AZI1 in vesicle‐like structures that move rapidly back and forth between chloroplasts and cytoplasm. Plastids showed elevated AZI1 levels during SAR, and move in close association with endoplasmic reticulum transcytoplasmic strands connecting chloroplasts, endoplasmic reticulum, and plasma membrane (Cecchini et al., 2015). Chloroplasts serve as synthesis sites of putative lipid signals (Cecchini et al., 2015). Observed AZI1 movement patterns suggest that it facilitates transport of SAR signals synthesized in chloroplasts (Figure 2).
Further studies showed that azelaic acid and G3P transport occurs via plasmodesmata and is regulated by plasmodesmata‐localizing proteins (PDLP) 1 and 5 (Lim et al., 2016). PDLP1 interacts with AZI1 but not with DIR1. These findings indicate that AZI1 transports SAR signals synthesized in chloroplasts to plasmodesmata via membrane vesicles, and DIR1 (or DIR1/AZI1) complexed with lipid signals is subsequently transported to systemic sites via plasmodesmata (Figure 2). This concept is consistent with the ability of DIR1 to move through phloem (Chanda et al., 2011), and the enrichment of AZI1 in phloem sap during SAR (Pitzschke et al., 2016). Recent studies show that AZI1 function in SAR is regulated by phosphorylation/dephosphorylation. Mitogen‐Activated Protein Kinase 3 and 6 (MPK3/6) promote AZI1 accumulation in plastids during priming induction and are functionally involved in systemic immunity (Cecchini et al., 2021; Pitzschke et al., 2014; Figure 2).
Besides transporting SAR activators via the pholem, AZI1 also is involved in defensive signal propagation between neighbouring plants. Riedlmeier et al. (2017) identified SAR‐associated volatile organic compounds emitted from Arabidopsis rosettes in response to P. syringae AvrRpm1. These compounds (e.g., α‐ and β‐pinene), when volatilized, induce defensive responses in neighbouring plants, including accumulation of reactive oxygen species and expression of SA‐ and SAR‐related genes. Such defensive signals do not increase in azi1 mutants however, suggesting that AZI1 mediates monoterpene‐induced defensive signal propagation. AZI1 also plays a role in transmitting systemic stomatal density response signals, which can inhibit pathogens by reducing stomatal density in systemic leaves (Dutton et al., 2019). See Figure 2 for an overview.
Protein–protein interactions play key roles in the transmission of stress signals. As mentioned above, DIR1 and AZI1 have important signalling functions in plant systemic immune processes. Further identification of AZI1‐ and DIR1‐interacting proteins is therefore necessary to understand their function in plant systemic immune responses. For example, DIR1 is translocated from pathogen‐inoculated tissues to systemic tissues via plasmodesmata (Lim et al., 2016). Identification of the receptor protein for DIR1 in systemic tissues would help to clarify the transmission mechanism of SAR signals. Two useful methods for identifying LTP‐interacting proteins are the yeast two‐hybrid system and co‐IP coupled with high‐resolution mass spectrometry. AZI1 and DIR1 also have been confirmed to transport the putative lipid SAR long‐distance signaling (Chanda et al., 2011; Maldonado et al., 2002). Thus, identification of lipids specifically bound by AZI1 and DIR1 during SAR induction is useful for identifying new SAR long‐distance signals. In the yeast Saccharomyces cerevisiae, the binding lipids of 13 LTPs were identified by co‐IP coupled with metabolomics based on high‐resolution mass spectrometry (Maeda et al., 2013). This method could also be used to identify bonding lipids of AZI1, DIR1 and other LTPs.
5. ROLE OF LTPs IN SIGNAL EXCHANGE BETWEEN HOST PLANT AND PARASITIC PATHOGENS
Leguminous plants are characterized by the ability to interact with rhizobia and produce nitrogen‐fixing organs (nodules). Medicago truncatula N5 (MtN5), a root‐specific LTP in the legume M. truncatula, is involved in symbiotic interaction between M. truncatula and Ensifer meliloti (formerly Sinorhizobium meliloti), and its expression is induced during such interaction (Pii et al., 2009). MtN5 production is induced in the early stage of E. meliloti infection and localized to mature nodules. The number of nodules produced in response to E. meliloti infection is higher in MtN5‐overexpressing lines and lower in MtN5‐knockdown lines. MtN5 displays lipid‐binding activity, and recombinant MtN5 binds lyso‐phosphatidylcholine in vivo (Pii et al., 2009). Phospholipase D, a key protein in arbuscular mycorrhizal symbiosis (Charron et al., 2004; Drissner et al., 2007), is functionally involved in MtN5 induction in E. meliloti‐inoculated roots (Pii et al., 2012). Together, these findings indicate LTP involvement in the signal exchange between host plant and rhizobia.
LTP AsE246, similar to MtN5, is required for nodule organogenesis in Astragalus sinicus (Chinese milk vetch) (Lei et al., 2014). AsE246 is expressed specifically in nodules and binds the plant‐synthesized membrane lipid digalactosyldiacylglycerol (DGDG) in vivo. AsE246 and DGDG are colocalized in the symbiosomal membrane. RNAi silencing of AsE246 expression in A. sinicus leads to reductions in the levels of phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol and DGDG. AsE246 knockdown results in decreased lipid contents in nodules, fewer nodule primordia and mature nodules, and fewer infection threads (Lei et al., 2014), suggesting that it assists lipid transport across the symbiosomal membrane and is essential for effective legume‐rhizobium symbiosis in A. sinicus. The high‐temperature protein G (HtpG) in Mesorhizobium huakuii interacts with AsE246 during legume‐rhizobium symbiosis and affects the symbiosomal lipid content in root nodules. It also assists in nodule development and nitrogen fixation (Zhou et al., 2019).
Most vascular plants benefit from symbiosis with mutualistic arbuscular mycorrhizal fungi (AMF), which facilitate nutrient and water uptake (Jiang et al., 2017). In return for mineral nutrients, plants transfer fixed carbon to AMF. Recent studies report that fatty acids synthesized in the host plants also are delivered to AMF and play essential roles in sustaining mycorrhizal colonization (Bravo et al., 2017; Luginbuehl et al., 2017; Rich et al., 2021). Fatty acids transfer depends on the ATP binding cassette in the transporter‐mediated lipid export pathway (Jiang et al., 2017). Furthermore, the symbiotic transfer of lipids in bryophytes is regulated by orthologous genetic pathways, similar to vascular plants, indicating its conservation across land plants (Rich et al., 2021). Based on their lipid transfer function, it is thus reasonable to speculate that LTPs also would play important roles in mycorrhizal colonization by transferring lipids from host plants to AMF.
6. LTPs MEDIATE SIGNAL TRANSMISSION IN ABSCISIC ACID SIGNALLING PATHWAY
Stomatal movement is regulated by various phytohormones, including SA, abscisic acid (ABA), and JA (Dutton et al., 2019, Xiang et al., 2021). ABA in particular plays a crucial role in controlling stomatal movements (Huang et al., 2021). Its content is elevated at pathogen infection sites to mediate closure of stomata and prevent further pathogen entry (David et al., 2019; Ding et al., 2016). StLTP10, a potato LTP, plays a key role in regulating Phytophthora infestans‐triggered stomatal closure by interacting with Pyrabatin Resistance 1‐Like 4 PYL4, an ABA receptor (Wang et al., 2021). StLTP10 biosynthesis is induced by P. infestans and phytohormones SA, ABA, and methyl salicylate, and StLTP10 overexpression enhances resistance to P. infestans in potato. Yeast two‐hybrid and bimolecular fluorescence complementation assays have confirmed this StLTP10–PYL4 interaction (Wang et al., 2021). StLTP10 recruits PYL4 to plant cell membranes and acts synergistically with PYL4 to induce stomatal closure in response to P. infestans infection (Wang et al., 2021) (Figure 3). Like StLTP10, another study similarly reports that AtLTP3 positively regulates ABA biosynthesis in Arabidopsis, and ABA levels increase in AtLTP3‐overexpressing plants (Gao et al., 2016).
FIGURE 3.
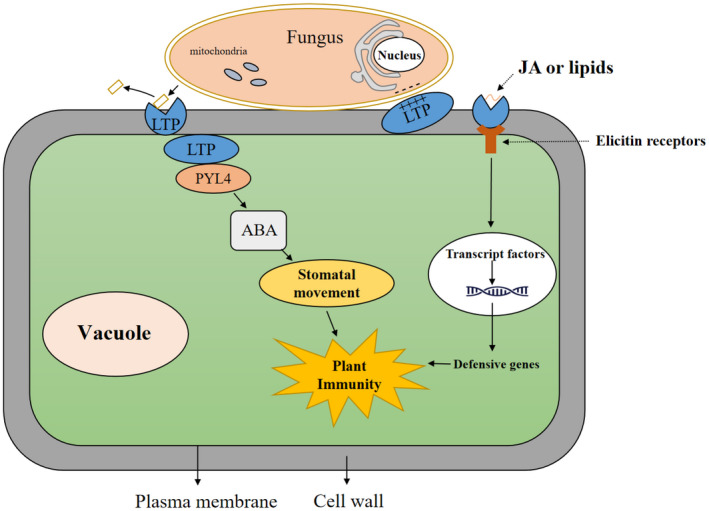
Lipid transfer protein (LTP) roles in plant and fungus interactions. LTPs can destroy fungal cell membranes using lipid transfer or highly conserved cationic residues. LTPs also regulate abscisic acid (ABA)‐mediated stomatal closure by interacting with abscisic acid receptor PYL4 to enhance resistance to fungi. Finally, LTP–jasmonic acid (JA) complexes bind competitively to elicitin receptors to activate immune responses
7. LTPs BIND COMPETITIVELY TO ELICITIN RECEPTORS TO ACTIVATE PLANT IMMUNE RESPONSES
Elicitins are small (c.10 kDa) cysteine‐enriched proteins secreted by pathogenic species of Phytophthora and Pythium, two genera of fungus‐like oomycetes (Buhot et al., 2001; Ponchet et al., 1999). Elicitins contain a hydrophobic cavity formed by an α‐helix fold and stabilized by three disulphide bonds (Boissy et al., 1999). This cavity allows the binding of sterol and fatty acids. In tobacco plants, sterol‐elicitin complexes recognize plasma membrane receptors and induce a hypersensitive response and SAR (Ponchet et al., 1999). Elicitins and plant LTPs have some common structural and nonspecific extracellular lipid‐binding properties. For example, the interaction of wheat LTP1 with elicitin receptors in tobacco plasma membranes indicates functional similarity between LTPs and elicitins (Buhot et al., 2001). The same group reported that tobacco LTP1 bound to various lipids, including the signalling molecule JA, in vivo (Buhot et al., 2004). LTP1–JA complexes bind to sites characterized as elicitin receptors, and exogenous localized application of the complexes in tobacco plants induces SAR against Phytophthora parasitica. In grape (Vitis vinifera), VvLTP4 interacts with JA, and exogenous application of VvLTP4–JA complex promotes resistance to B. cinerea (Girault et al., 2008). JA also plays a key functional role in plant defence responses to necrotrophic pathogens (Glazebrook, 2005). These findings demonstrate the ability of plant LTPs to bind elicitor receptors and mediate pathogen resistance via the JA signalling pathway (Figure 3).
8. ROLE OF LTP IN NEGATIVE REGULATION OF PLANT IMMUNE RESPONSES
Plant LTPs most commonly function in plant immune processes as positive factors. As members of the PR14 family of PR proteins, LTPs show inducible expression patterns during pathogen challenge (Sels et al., 2008). However, some LTPs negatively regulate plant immune responses. During SAR in Arabidopsis, DIR1 transfers a putative lipid signal (Chanda et al., 2011; Maldonado et al., 2002). Heterologous expression of AtDIR1, OsDIR1‐A, and OsDIR1‐B in barley promotes local defensive responses (Colebrook, 2010). However, TaDIR1‐2, the wheat ortholog of DIR1, acts as a negative regulator in wheat resistance to Puccinia striiformis f. sp. tritici by modulating reactive oxygen species (ROS) and SA‐induced signalling (Ahmed et al., 2017). Specifically, taDIR1‐2‐knockdown plants displayed accumulation of ROS and SA, and enhanced resistance to P. striiformis. In Arabidopsis, AtLTP3 similarly contributes to disease susceptibility by enhancing ABA biosynthesis (Gao et al., 2016). LTP3‐overexpressing strains had increased ABA levels, reduced SA levels, and increased susceptibility to Pseudomonas syringae (Gao et al., 2016). Previous studies indicated that ABA acts antagonistically against SA‐mediated immune signalling (David et al., 2019). However, another study indicated that full‐scale induction of SA and NPR1‐dependent genes requires ABA signalling (Ding et al., 2016). Although both regulate cellular NPR1 protein levels in an antagonistic manner, ABA is critical for SA accumulation and induction of SA‐dependent defence responses in both P. syringae‐infected tissues and adjacent tissues (Ding et al., 2016). The accumulation level of ABA increased in both P. syringae‐infected tissues and adjacent tissues, and the induction of SA and SA‐dependent genes was compromised in aba3 plants defective in ABA biosynthesis during P. syringae infection (Ding et al., 2016). These findings indicate that dynamic homeostasis of ABA and SA during plant immune processes is essential for full‐scale induction of defence responses. Thus, these LTPs, which appear to negatively regulate plant immunity, may be critical for the induction of an intact resistance response by modifying the ABA/SA balance (Figure 1). Further elucidation of the mechanisms by which these LTPs regulate phytohormone homeostasis will greatly improve our understanding of the molecular mechanisms of plant immunity.
Like TaDIR1‐2 and AtLTP3, Accelerated Cell Death 11 (ACD11) in Arabidopsis also has a negative effect on plant immunity (Brodersen et al., 2002). ACD11 encodes an LTP and is able to transfer ceramide‐1‐phosphate (C1P), which plays important roles in plant growth, development, senescence and programmed cell death (PCD) (Gao et al., 2022; Simanshu et al., 2014; Zhai et al., 2017). X‐ray structures showed that ACD11 belongs to the glycolipid transfer protein superfamily. ACD11 contains a surface‐localized, phosphate headgroup recognition centre connected to an interior hydrophobic pocket, which can selectively bind and transfer C1P (Simanshu et al., 2014). Knockout of ACD11 results in activation of PCD and the SA‐dependent defence response (Brodersen et al., 2002, 2005). The phenotype of acd11 plants occurs due to ectopic activation of LAZ (Lazarus) 1, LAZ2, LAZ4 and LAZ5 (Malinovsky et al., 2010; Munch et al., 2015; Palma et al., 2010). In Arabidopsis, the binding partner of ACD11 and its homologs also negatively regulate PCD and the defence response by interacting with ACD11 (Li et al., 2019; Petersen et al., 2009). Recently, a RING‐type E3 ligase, XBAT35.2, was found to positively regulate defence responses against Pst DC3000 by promoting the ubiquitination and subsequent degradation of ACD11 in Arabidopsis (Liu et al., 2017). A low accumulation of XBAT35.2 is maintained via self‐regulation under normal growth conditions, and ACD11 thus could accumulate and suppress PCD. In the presence of Pst DC3000, however, XBAT35.2 levels increase to promote ACD11 degradation, allowing for PCD and expression of SA‐dependent genes to occur as part of the defence response (Liu et al., 2017).
Higher plants have evolved multiple immune systems to restrict colonization and invasion (Pastorczyk‐Szlenkier & Bednarek, 2021). However, constitutive activation of immune responses, such as PAMP‐triggered immunity, effector‐triggered immunity and SAR, will allocate nutrient resources into the biosynthesis of multiple defence molecules, which in turn often inhibit plant growth and development (Pastorczyk‐Szlenkier & Bednarek, 2021). Thus, ACD11 may serve a essential role in balancing plant growth and immunity: under normal conditions, it helps to maintain normal growth and development by suppressing PCD (Liu et al., 2017), while under biotic stress XBAT35.2 accumulation promotes ACD11 degradation to initiate PCD and defence responses (Liu et al., 2017).
9. CONCLUSION
Plant LTPs play a variety of roles in plant immune responses. Although no single function can be assigned to all LTPs in general, individual isoforms play specific and sometimes multiple biological roles, such as directly inhibiting fungal and bacterial growth, regulating the cell wall structure, signalling transduction during SAR, nodule formation and participating in phytohormone signalling pathways (Table 1). Several questions remain, however, regarding the precise mechanisms of LTPs involved in plant–pathogen interactions. For example, can LTPs inhibit the growth of pathogens by transporting lipids synthesized in plant cells into pathogens? Do LTPs play a role in mycorrhizal colonization by transporting fatty acids from host cells to symbiotic pathogens? How do LTPs regulate plant defence responses by regulating phytohormone homeostasis? Can LTPs directly bind to and inhibit constituent proteins of the bacterial secretion system? How do LTPs regulate the plant growth–immunity trade‐off in plant–pathogen interactions? Finally, if lipid‐binding activity is necessary for the defensive functions of LTPs, which lipids are bound by LTPs in plant–pathogen interactions? Clarifying these aspects will greatly advance the understanding of LTP functions.
TABLE 1.
Summary of lipid transfer proteins identified in plants and their functions
| Species | Protein name | Functions | References |
|---|---|---|---|
| Arabidopsis thaliana | SCP‐2 | Edqvist et al. (2004) | |
| LTP6 | Chae et al. (2010) | ||
| LTP5 | Pollen tube development | Chae et al. (2009, 2010) | |
| DIR1 | Systemic acquired resistance | Chanda et al. (2011), Maldonado et al. (2002) | |
| DIR1‐like | Systemic acquired resistance | Champigny et al. (2013) | |
| LTPg3, LTPg4 | Pollen development | Edstam and Edqvist (2014) | |
| LTP3 | Germination and seedling growth, freezing and drought stress | Guo, Yang, et al. (2013), Pagnussat et al. (2015) | |
| END1 | Li, Lopato, et al. (2014) | ||
| AtLTP4.5 | Biotic stress | McLaughlin et al. (2015) | |
| EARLI1 | Systemic acquired resistance | Cecchini et al. (2015) | |
| AZI1 | Systemic acquired resistance, salt stress | Cecchini et al. (2015), Jung et al. (2009), Pitzschke et al. (2014) | |
| LTPg15‐LTPg17, LTPg20, LTPg22, LTPg23, LTPg26, LTPg30 | Edstam et al. (2013) | ||
| LTP1 | Ethylene‐mediated signalling pathway, cell differentiation, embryo and shoot development | Baroux et al. (2001), Potocka et al. (2012), Toonen et al. (1997), Wang et al. (2016), | |
| AtLtpI‐4 | Suberin formation of crown galls | Deeken et al. (2016) | |
| ACD11 | Negative regulation of programmed cell death | Brodersen et al. (2002, 2005), Simanshu et al. (2014), Zhai et al. (2017) | |
| AtLTP2 | Maintaining the integrity of cell wall | Jacq et al. (2017) | |
| LTPg15 | Seed coat permeability | Lee and Suh (2018) | |
| LTPg6 | Defence responses to fungi | Fahlberg et al. (2019) | |
| LTPg2 | Defence responses to fungal cuticular wax export | Fahlberg et al. (2019), Kim et al. (2012) | |
| LTPg1 | Defence responses to fungal cuticular wax export | Debono et al. (2009), Fahlberg et al. (2019), Kim et al. (2012), Lee et al. (2009) | |
| LTPg5 | Defence responses to bacteria and fungi, seed development | Ali et al. (2020), Edstam and Edqvist (2014), Fahlberg et al. (2019) | |
| DRN1 | Defence responses, salt stress | Dhar et al. (2020) | |
| AtLTP4.4 | Antifungal, antioxidant | McLaughlin et al. (2021) | |
| LSR1 | Regulate leaf senescence | Feng et al. (2022) | |
| Oryza sativa | LTP | Lee et al. (1998) | |
| LTP‐2 | Samue et al. (2002) | ||
| OsLTP5 | Defence responses | Kim et al. (2008) | |
| OsDIL | Drought stress, development | Guo, Ge, et al. (2013) | |
| Psd1 | Growth and development | Li, Xia, et al. (2014) | |
| OsLTPL36 | Seed development and germination | Wang et al. (2015) | |
| Ptd1 | Growth and development | Deng et al. (2020) | |
| OsLTPL159 | Cold tolerance | Zhao et al. (2020) | |
| OsLTPL94 | Pollen wall development | Tao et al. (2021) | |
| OsC6 | Pollen wall development, anther development | Chen et al. (2022), Zhang et al. (2010) | |
| OsLTP47 | Pollen wall development | Chen et al. (2022) | |
| Triticum aestivum | LTP1 | Charvolin et al. (1999), Gincel et al. (1994) | |
| AceAMP1 | Defence responses, antifungal | Roy‐Barman et al. (2006) | |
| TaLtp9.1b, TaLTP9.2b‐TaLTP9.2d, TaLtp9.3a‐TaLtp9.3g, TaLtp9.4a‐TaLtp9.4c, TaLtp9.7a‐TaLtp9.7e, TaLTP7.1a‐TaLTP7.1c | Boutrot et al. (2007, 2008) | ||
| TaLt10B6, TaBs108F7, TaLt10F9 | Antifungal | Sun et al. (2008) | |
| Ltp3F1 | Antifungal | Kirubakaran et al. (2008) | |
| TaPR60 | Binding of lipid molecules | Kovalchuk et al. (2009) | |
| TaLTP5 | Defence responses against fungi | Zhu et al. (2012) | |
| TaPR61 | Kovalchuk et al. (2009) | ||
| TaDIR1‐2 | Negative regulator in wheat resistance to fungi | Ahmed et al. (2017) | |
| Ms1 | Pollen exine development | Tucker et al. (2017) | |
| TaLTP40, TaLTP75 | Salt tolerance | Hairat et al. (2018) | |
| TaMs1 | Pollen development | Kouidri et al. (2018) | |
| TaMs5‐A, TaMs5‐B | Pollen exine development | Pallotta et al. (2019) | |
| TaLTP3 | Defence responses against Puccinia triticina, thermotolerance and oxidative stress | Wang et al. (2017), Zhao et al. (2021) | |
| Euphorbia lagascae | ElLTP1, EILT2P2 | Programmed cell death | Edqvist and Farbos (2002), Eklund and Edqvist (2003) |
| Malus pumila | LTP3 | Cuticle formation | André et al. (2022) |
| Triticum durum | TdPR61 | Kovalchuk et al. (2012) | |
| TdLTP4 | Antimicrobial, abiotic and biotic stress | Ben et al. (2021), Safi et al. (2015) | |
| Pisum sativum | PsLTP1 | Binding abscisic acid | Akhiyarova et al. (2021) |
| Artemisia annua | AaLTP3, AaLTP4 | Growth and development | Adhikari et al. (2019) |
| Gossypium hirsutum | GhLTPg1 | Fibre elongation | Deng et al. (2016) |
| GhnsLTPsA10 | Defence responses against Verticillium wilt | Chen et al. (2021) | |
| Bassica napus | BnLTP‐II | Defence responses against Pseudomonas syringe pv. tomato | Balmant et al. (2021) |
| BraLTP1 | Epicuticular wax deposition and development | Liu et al. (2014) | |
| BraLTP2 | Trichome development | Tian et al. (2018) | |
| Chrysanthemum morifolium | DgnsLTP | Cold tolerance | Huang et al. (2021) |
| Solanum tuberosum | StnsLTP1 | Aiotic stresses | Gangadhar et al. (2016) |
| StLTP10 | Defence responses against Phytophthora infestans | Wang et al. (2021) | |
| Trachyspermum ammi | nsLTP1 | Nazeer et al. (2019) | |
| Zea mays | BETL9, BETL9‐like | Royo et al. (2014) | |
| Ms44 | Dominant male sterility | Fox et al. (2017) | |
| Nicotiana benthamiana | NbLTP1 | Assists bamboo mosaic virus Bamboo mosaic virus accumulation | Chiu et al. (2020) |
| Nicotiana tabacum | TobLTP2 | Cell wall extension | Nieuwland et al. (2005) |
| NtLTP1‐NtLTP4 | Promote monoterpene emission, lipid secretion from glandular trichomes, salt and drought stresses | Choi et al. (2012), Hwang et al. (2020), Xu et al. (2018) | |
| Triticosecale | LTPc3a, LTPc3b | Pollen wall development | Zaidi et al. (2020) |
| Brassica rapa | BrLTP2.1 | Antifungal | Schmitt et al. (2018) |
| Morinda citrifolia | McLTP1 | Antibacterial | Souza et al. (2018) |
| Medicago truncatula | MtN5 | Rhizobium–host interaction | Pii et al. (2009,2012) |
| Chelidonium majus | CmLTP9.5 | Antibacterial | Nawrot et al. (2017) |
| Setaria italica | SiLTP | Salt and drought tolerance | Pan et al. (2016) |
| Coffea canephora | CcLTP2 | Antimicrobial activity against pathogens | Bard et al. (2016) |
| Coffea arabica | CaLTP1a, CaLTP1b, CaLTP3a, CaLTP3b | Cotta et al. (2014) | |
| Panax ginseng | pgLTP | Antifungal | Cai et al. (2016) |
| Cucumis sativus | CsDIR1, CsDIR2 | Systemic acquired resistance | Isaacs et al. (2016) |
| Lotus japonicus | LjLTP10 | Drought stress, cutin formation | Tapia et al. (2013) |
| Astragalus sinicus | AsE246 | Legume‐rhizobium symbiosis | Lei et al. (2014) |
| Lens culinaris | LcLTP2 | Antimicrobia | Gizatullina et al. (2013) |
| Helianthus annuus | HaAP10 | Seed germination | Pagnussat et al. (2012) |
| Capsicum annuum | CaLTP(1) | Antifungal | Diz et al. (2011) |
| CaMF2 | Pollen development | Chen et al. (2011) | |
| CALTPI, CALTPII | Local and systemic acquired resistance | Sarowar et al. (2009) | |
| Leonurus japonicus | LJAMP2 | Defence responses against fungi | Jia et al. (2010) |
| Vitis vinifera | VvLTP2‐VvLTP5 | Defence responses against fungi | Girault et al. (2008) |
| VvLTP1 | Embryo development | François et al. (2008) | |
| Sesamum indicum | SiLTP1‐SiLTP5 | Choi et al. (2008) | |
| Senecia squalidus | SsLTP1 | Osmotic constraints, cold acclimation | Kielbowicz‐Matuk et al. (2008) |
| Vigna radiata | Vrltp1, Vrltp2 | Liu and Lin (2003) | |
| Mb‐nsLTP1 | Lin et al. (2005) | ||
| Physcomitrella patens | PpLTPg2, PpLTPG8 | Drought and cold stress | Edstam and Edqvist (2014) |
| Hordeum vulgare | HvLTP1.1‐HvLTP1.16, HvLTP2.1‐HvLTP2.5, HvLTPd1‐HvLTPd11, HvLTPg1‐HvLTPg8 | Duo et al. (2021) | |
| Ginkgo biloba | Gb‐nsLTP1 | Sawano et al. (2008) |
ACKNOWLEDGEMENTS
This work was supported by the Doctoral Research Start‐up Fund Project of Shangqiu Normal University (7001700235). The authors are grateful to Dr S. Anderson for English editing of the manuscript.
Gao, H. , Ma, K. , Ji, G. , Pan, L. & Zhou, Q. (2022) Lipid transfer proteins involved in plant–pathogen interactions and their molecular mechanisms. Molecular Plant Pathology, 23, 1815–1829. Available from: 10.1111/mpp.13264
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Adhikari, P.B. , Han, J.Y. , Ahn, C.H. & Choi, Y.E. (2019) Lipid transfer proteins (AaLTP3 and AaLTP4) are involved in sesquiterpene lactone secretion from glandular trichomes in Artemisia annua . Plant & Cell Physiology, 60, 2826–2836. [DOI] [PubMed] [Google Scholar]
- Ahmed, S.M. , Liu, P. , Xue, Q. , Ji, C. , Qi, T. , Guo, J. et al. (2017) TaDIR1‐2, a wheat ortholog of lipid transfer protein AtDIR1 contributes to negative regulation of wheat resistance against Puccinia striiformis f. sp. tritici . Frontiers in Plant Science, 8, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhiyarova, G.R. , Ivanov, R.S. , Ivanov, I.I. , Finkina, E.I. , Melnikova, D.N. , Bogdanov, I.V. et al. (2021) Effects of salinity and abscisic acid on lipid transfer protein accumulation, suberin deposition and hydraulic conductance in pea roots. Membranes, 11, 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, M.A. , Abbas, A. , Azeem, F. , Shahzadi, M. & Bohlmann, H. (2020) The Arabidopsis GPI‐anchored LTPg5 encoded by At3g22600 has a role in resistance against a diverse range of pathogens. International Journal of Molecular Sciences, 21, 1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre, C.M. , Guerriero, G. , Lateur, M. , Charton, S. , Leclercq, C.C. , Renaut, J. et al. (2022) Identification of novel candidate genes involved in apple cuticle integrity and russeting‐associated triterpene synthesis using metabolomic, proteomic, and transcriptomic data. Plants, 11, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmant, K.M. , Lawrence, S.N. , Duong, B.V. , Zhu, F. , Zhu, N. , Nicklay, J. et al. (2021) Guard cell redox proteomics reveals a role of lipid transfer protein in plant defense. Journal of Proteomics, 242, 104247. [DOI] [PubMed] [Google Scholar]
- Bard, G.C. , Zottich, U. , Souza, T.A. , Ribeiro, S.F. , Dias, G.B. , Pireda, S. et al. (2016) Purification, biochemical characterization, and antimicrobial activity of a new lipid transfer protein from Coffea canephora seeds. Genetic & Molecular Research, 15, 1–6. [DOI] [PubMed] [Google Scholar]
- Baroux, C. , Blanvillain, R. , Moore, I.R. & Gallois, P. (2001) Transactivation of BARNASE under the AtLTP1 promoter affects the basal pole of the embryo and shoot development of the adult plant in Arabidopsis . The Plant Journal, 28, 503–515. [DOI] [PubMed] [Google Scholar]
- Ben, H.A. , Ben, S.R. , Dhifi, W. , Mnif, W. & Brini, F. (2021) Novel non‐specific lipid‐transfer protein (TdLTP4) isolated from durum wheat: antimicrobial activities and anti‐inflammatory properties in lipopolysaccharide (LPS)‐stimulated RAW 264.7 macrophages. Microbial Pathogenesis, 154, 104869. [DOI] [PubMed] [Google Scholar]
- Boissy, G. , O'Donohue, M. , Gaudemer, O. , Perez, V. , Pernollet, J.C. & Brunie, S. (1999) The 2.1 A structure of an elicitin‐ergosterol complex: a recent addition to the Sterol Carrier Protein family. Protein Science, 8, 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot, F. , Chantret, N. & Gautier, M.F. (2008) Genome‐wide analysis of the rice and Arabidopsis non‐specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics, 9, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot, F. , Meynard, D. , Guiderdoni, E. , Joudrier, P. & Gautier, M.F. (2007) The Triticum aestivum non‐specific lipid transfer protein (TaLtp) gene family: comparative promoter activity of six TaLtp genes in transgenic rice. Planta, 225, 843–862. [DOI] [PubMed] [Google Scholar]
- Bravo, A. , Brands, M. , Wewer, V. , Dormann, P. & Harrison, M.J. (2017) Arbuscular mycorrhiza‐specific enzymes FatM and RAM2 fine‐tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Pphytologist, 214, 1631–1645. [DOI] [PubMed] [Google Scholar]
- Brodersen, P. , Malinovsky, F.G. , Hematy, K. , Newman, M.A. & Mundy, J. (2005) The role of salicylic acid in the induction of cell death in Arabidopsis acd11 . Plant Physiology, 138, 1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P. , Petersen, M. , Pike, H.M. , Olszak, B. , Skov, S. , Odum, N. et al. (2002) Knockout of Arabidopsis accelerated‐cell‐death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes & Development, 16, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhot, N. , Douliez, J.P. , Jacquemard, A. , Marion, D. , Tran, V. , Maume, B.F. et al. (2001) A lipid transfer protein binds to a receptor involved in the control of plant defence responses. FEBS Letters, 509, 27–30. [DOI] [PubMed] [Google Scholar]
- Buhot, N. , Gomes, E. , Milat, M.L. , Ponchet, M. , Marion, D. , Lequeu, J. et al. (2004) Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Molecular Biology of the Cell, 15, 5047–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, K. , Wang, J. , Wang, M. , Zhang, H. , Wang, S. & Zhao, Y. (2016) Molecular cloning, recombinant expression, and antifungal functional characterization of the lipid transfer protein from Panax ginseng . Biotechnology Letters, 38, 1229–1235. [DOI] [PubMed] [Google Scholar]
- Cameron, K.D. , Teece, M.A. & Smart, L.B. (2006) Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiology, 140, 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini, N.M. , Speed, D.J. , Roychoudhry, S. & Greenberg, J.T. (2021) Kinases and protein motifs required for AZI1 plastid localization and trafficking during plant defense induction. The Plant Journal, 105, 1615–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini, N.M. , Steffes, K. , Schlappi, M.R. , Gifford, A.N. & Greenberg, J.T. (2015) Arabidopsis AZI1 family proteins mediate signal mobilization for systemic defence priming. Nature Communications, 6, 7658. [DOI] [PubMed] [Google Scholar]
- Chae, K. , Gonong, B.J. , Kim, S.C. , Kieslich, C.A. , Morikis, D. , Balasubramanian, S. et al. (2010) A multifaceted study of stigma/style cysteine‐rich adhesin (SCA)‐like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. Journal of Experimental Botany, 61, 4277–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, K. , Kieslich, C.A. , Morikis, D. , Kim, S.C. & Lord, E.M. (2009) A gain‐of‐function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. The Plant Cell, 21, 3902–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champigny, M.J. , Isaacs, M. , Carella, P. , Faubert, J. , Fobert, P.R. & Cameron, R.K. (2013) Long distance movement of DIR1 and investigation of the role of DIR1‐like during systemic acquired resistance in Arabidopsis . Frontiers in Plant Science, 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda, B. , Xia, Y. , Mandal, M.K. , Yu, K. , Sekine, K.T. , Gao, Q.M. et al. (2011) Glycerol‐3‐phosphate is a critical mobile inducer of systemic immunity in plants. Nature Genetics, 43, 421–427. [DOI] [PubMed] [Google Scholar]
- Charron, D. , Pingret, J.L. , Chabaud, M. , Journet, E.P. & Barker, D.G. (2004) Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiology, 136, 3582–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvolin, D. , Douliez, J.P. , Marion, D. , Cohen‐Addad, C. & Pebay‐Peyroula, E. (1999) The crystal structure of a wheat nonspecific lipid transfer protein (ns‐LTP1) complexed with two molecules of phospholipid at 2.1 A resolution. European Journal of Biochemistry, 264, 562–568. [DOI] [PubMed] [Google Scholar]
- Chassot, C. , Nawrath, C. & Metraux, J.P. (2007) Cuticular defects lead to full immunity to a major plant pathogen. The Plant Journal, 49, 972–980. [DOI] [PubMed] [Google Scholar]
- Chaturvedi, R. , Venables, B. , Petros, R.A. , Nalam, V. , Li, M. , Wang, X. et al. (2012) An abietane diterpenoid is a potent activator of systemic acquired resistance. The Plant Journal, 71, 161–172. [DOI] [PubMed] [Google Scholar]
- Chen, B. , Zhang, Y. , Sun, Z. , Liu, Z. , Zhang, D. , Yang, J. et al. (2021) Tissue‐specific expression of GhnsLTPs identified via GWAS sophisticatedly coordinates disease and insect resistance by regulating metabolic flux redirection in cotton. The Plant Journal, 107, 831–846. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Chen, G. , Hao, X. , Cao, B. , Chen, Q. , Liu, S. et al. (2011) CaMF2, an anther‐specific lipid transfer protein (LTP) gene, affects pollen development in Capsicum annuum L. Plant Science, 181, 439–448. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Ji, C. , Zhou, D. , Gou, X. , Tang, J. , Jiang, Y. et al. (2022) OsLTP47 may function in a lipid transfer relay essential for pollen wall development in rice. Journal of Genetics and Genomics, 49, 481–491. [DOI] [PubMed] [Google Scholar]
- Chiu, L.Y. , Chen, I.H. , Hsu, Y.H. & Tsai, C.H. (2020) The lipid transfer protein 1 from Nicotiana benthamiana assists Bamboo mosaic virus accumulation. Viruses, 12, 1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, A.M. , Lee, S.B. , Cho, S.H. , Hwang, I. , Hur, C.G. & Suh, M.C. (2008) Isolation and characterization of multiple abundant lipid transfer protein isoforms in developing sesame (Sesamum indicum L.) seeds. Plant Physiology and Biochemistry, 46, 127–139. [DOI] [PubMed] [Google Scholar]
- Choi, Y.E. , Lim, S. , Kim, H.J. , Han, J.Y. , Lee, M.H. , Yang, Y. et al. (2012) Tobacco NtLTP1, a glandular‐specific lipid transfer protein, is required for lipid secretion from glandular trichomes. The Plant Journal, 70, 480–491. [DOI] [PubMed] [Google Scholar]
- Colebrook, E. (2010) The localisation of Pseudomonas‐induced acquired resistance in barley. PhD thesis. Norwich: University of East Anglia. [Google Scholar]
- Cotta, M.G. , Barros, L.M. , de Almeida, J.D. , de Lamotte, F. , Barbosa, E.A. , Vieira, N.G. et al. (2014) Lipid transfer proteins in coffee: isolation of Coffea orthologs, Coffea arabica homeologs, expression during coffee fruit development and promoter analysis in transgenic tobacco plants. Plant Molecular Biology, 85, 11–31. [DOI] [PubMed] [Google Scholar]
- David, L. , Harmon, A.C. & Chen, S. (2019) Plant immune responses – from guard cells and local responses to systemic defense against bacterial pathogens. Plant Signaling & Behavior, 14, e1588667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Samblanx, G.W. , Goderis, I.J. , Thevissen, K. , Raemaekers, R. , Fant, F. , Borremans, F. et al. (1997) Mutational analysis of a plant defensin from radish (Raphanus sativus L.) reveals two adjacent sites important for antifungal activity. Journal of Biological Chemistry, 272, 1171–1179. [DOI] [PubMed] [Google Scholar]
- Debono, A. , Yeats, T.H. , Rose, J.K. , Bird, D. , Jetter, R. , Kunst, L. et al. (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol‐anchored lipid transfer protein required for export of lipids to the plant surface. The Plant Cell, 21, 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken, R. , Saupe, S. , Klinkenberg, J. , Riedel, M. , Leide, J. , Hedrich, R. et al. (2016) The nonspecific lipid transfer protein AtLtpI‐4 is involved in suberin formation of Arabidopsis thaliana crown galls. Plant Physiology, 172, 1911–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, T. , Yao, H. , Wang, J. , Wang, J. , Xue, H. & Zuo, K. (2016) GhLTPG1, a cotton GPI‐anchored lipid transfer protein, regulates the transport of phosphatidylinositol monophosphates and cotton fiber elongation. Scientific Reports, 6, 26829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W. , Li, R. , Xu, Y. , Mao, R. , Chen, S. , Chen, L. et al. (2020) A lipid transfer protein variant with a mutant eight‐cysteine motif causes photoperiod‐ and thermo‐sensitive dwarfism in rice. Journal of Experimental Botany, 71, 1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W. , Marshall, N.C. , Rowland, J.L. , McCoy, J.M. , Worrall, L.J. , Santos, A.S. et al. (2017) Assembly, structure, function and regulation of type III secretion systems. Nature Reviews Microbiology, 15, 323–337. [DOI] [PubMed] [Google Scholar]
- Dhar, N. , Caruana, J. , Erdem, I. & Raina, R. (2020) An Arabidopsis DISEASE RELATED NONSPECIFIC LIPID TRANSFER PROTEIN 1 is required for resistance against various phytopathogens and tolerance to salt stress. Gene, 753, 144802. [DOI] [PubMed] [Google Scholar]
- Ding, Y. , Dommel, M. & Mou, Z. (2016) Abscisic acid promotes proteasome‐mediated degradation of the transcription coactivator NPR1 in Arabidopsis thaliana . The Plant Journal, 86, 20–34. [DOI] [PubMed] [Google Scholar]
- Diz, M.S. , Carvalho, A.O. , Ribeiro, S.F. , Da, C.M. , Beltramini, L. , Rodrigues, R. et al. (2011) Characterisation, immunolocalisation and antifungal activity of a lipid transfer protein from chili pepper (Capsicum annuum) seeds with novel α‐amylase inhibitory properties. Physiologia Plantarum, 142, 233–246. [DOI] [PubMed] [Google Scholar]
- Drissner, D. , Kunze, G. , Callewaert, N. , Gehrig, P. , Tamasloukht, M. , Boller, T. et al. (2007) Lyso‐phosphatidylcholine is a signal in the arbuscular mycorrhizal symbiosis. Science, 318, 265–268. [DOI] [PubMed] [Google Scholar]
- Duo, J. , Xiong, H. , Wu, X. , Li, Y. , Si, J. , Zhang, C. et al. (2021) Genome‐wide identification and expression profile under abiotic stress of the barley non‐specific lipid transfer protein gene family and its Qingke orthologues. BMC Genomics, 22, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton, C. , Horak, H. , Hepworth, C. , Mitchell, A. , Ton, J. , Hunt, L. et al. (2019) Bacterial infection systemically suppresses stomatal density. Plant Cell & Environment, 42, 2411–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist, J. & Farbos, I. (2002) Characterization of germination‐specific lipid transfer proteins from Euphorbia lagascae . Planta, 215, 41–50. [DOI] [PubMed] [Google Scholar]
- Edqvist, J. , Ronnberg, E. , Rosenquist, S. , Blomqvist, K. , Viitanen, L. , Salminen, T.A. et al. (2004) Plants express a lipid transfer protein with high similarity to mammalian sterol carrier protein‐2. Journal of Biological Chemistry, 279, 53544–53553. [DOI] [PubMed] [Google Scholar]
- Edstam, M.M. & Edqvist, J. (2014) Involvement of GPI‐anchored lipid transfer proteins in the development of seed coats and pollen in Arabidopsis thaliana . Physiologia Plantarum, 152, 32–42. [DOI] [PubMed] [Google Scholar]
- Edstam, M.M. , Blomqvist, K. , Eklof, A. , Wennergren, U. & Edqvist, J. (2013) Coexpression patterns indicate that GPI‐anchored non‐specific lipid transfer proteins are involved in accumulation of cuticular wax, suberin and sporopollenin. Plant Molecular Biology, 83, 625–649. [DOI] [PubMed] [Google Scholar]
- Edstam, M.M. , Laurila, M. , Hoglund, A. , Raman, A. , Dahlstrom, K.M. , Salminen, T.A. et al. (2014) Characterization of the GPI‐anchored lipid transfer proteins in the moss Physcomitrella patens . Plant Physiology & Biochemistry, 75, 55–69. [DOI] [PubMed] [Google Scholar]
- Edstam, M.M. , Viitanen, L. , Salminen, T.A. & Edqvist, J. (2011) Evolutionary history of the non‐specific lipid transfer proteins. Molecular Plant, 4, 947–964. [DOI] [PubMed] [Google Scholar]
- Eklund, D.M. & Edqvist, J. (2003) Localization of nonspecific lipid transfer proteins correlate with programmed cell death responses during endosperm degradation in Euphorbia lagascae seedlings. Plant Physiology, 132, 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlberg, P. , Buhot, N. , Johansson, O.N. & Andersson, M.X. (2019) Involvement of lipid transfer proteins in resistance against a non‐host powdery mildew in Arabidopsis thaliana . Molecular Plant Pathology, 20, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, G. , Zhong, Y. & Zou, W. (2022) Lipid transporter LSR1 positively regulates leaf senescence in Arabidopsis . Plant Signaling & Behavior, 17, 2007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, T. , DeBruin, J. , Haug, C.K. , Trimnell, M. , Clapp, J. , Leonard, A. et al. (2017) A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnology Journal, 15, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois, J. , Lallemand, M. , Fleurat‐Lessard, P. , Laquitaine, L. , Delrot, S. , Coutos‐Thevenot, P. et al. (2008) Overexpression of the VvLTP1 gene interferes with somatic embryo development in grapevine. Functional Plant Biology, 35, 394–402. [DOI] [PubMed] [Google Scholar]
- Gangadhar, B.H. , Sajeesh, K. , Venkatesh, J. , Baskar, V. , Abhinandan, K. , Yu, J.W. et al. (2016) Enhanced tolerance of transgenic potato plants over‐expressing non‐specific lipid transfer protein‐1 (StnsLTP1) against multiple abiotic stresses. Frontiers in Plant Science, 7, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H. , Guo, M. , Song, J. , Ma, Y. & Xu, Z. (2021) Signals in systemic acquired resistance of plants against microbial pathogens. Molecular Biology Reports, 48, 3747–3759. [DOI] [PubMed] [Google Scholar]
- Gao, S. , Guo, W. , Feng, W. , Liu, L. , Song, X. , Chen, J. et al. (2016) LTP3 contributes to disease susceptibility in Arabidopsis by enhancing abscisic acid (ABA) biosynthesis. Molecular Plant Pathology, 17, 412–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y.G. , McDonald, J. , Malinina, L. , Patel, D.J. & Brown, R.E. (2022) Ceramide‐1‐phosphate transfer protein promotes sphingolipid reorientation needed for binding during membrane interaction. Journal of Lipid Research, 63, 100151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X. , Chen, J. , Sun, C. & Cao, K. (2003) Preliminary study on the structural basis of the antifungal activity of a rice lipid transfer protein. Protein Engineering, 16, 387–390. [DOI] [PubMed] [Google Scholar]
- Gincel, E. , Simorre, J.P. , Caille, A. , Marion, D. , Ptak, M. & Vovelle, F. (1994) Three‐dimensional structure in solution of a wheat lipid‐transfer protein from multidimensional 1H‐NMR data. A new folding for lipid carriers. European Journal of Biochemistry, 226, 413–422. [DOI] [PubMed] [Google Scholar]
- Girault, T. , Francois, J. , Rogniaux, H. , Pascal, S. , Delrot, S. , Coutos‐Thevenot, P. et al. (2008) Exogenous application of a lipid transfer protein‐jasmonic acid complex induces protection of grapevine towards infection by Botrytis cinerea . Plant Physiology and Biochemistry, 46, 140–149. [DOI] [PubMed] [Google Scholar]
- Gizatullina, A.K. , Finkina, E.I. , Mineev, K.S. , Melnikova, D.N. , Bogdanov, I.V. , Telezhinskaya, I.N. et al. (2013) Recombinant production and solution structure of lipid transfer protein from lentil Lens culinaris . Biochemical and Biophysical Research Communications, 439, 427–432. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology, 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Guo, C. , Ge, X. & Ma, H. (2013) The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Molecular Biology, 82, 239–253. [DOI] [PubMed] [Google Scholar]
- Guo, L. , Yang, H. , Zhang, X. & Yang, S. (2013) Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis . Journal of Experimental Botany, 64, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hairat, S. , Baranwal, V.K. & Khurana, P. (2018) Identification of Triticum aestivum nsLTPs and functional validation of two members in development and stress mitigation roles. Plant Physiology and Biochemistry, 130, 418–430. [DOI] [PubMed] [Google Scholar]
- Hoffmann‐Benning, S. & Kende, H. (1994) Cuticle biosynthesis in rapidly growing internodes of deepwater rice. Plant Physiology, 104, 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Q. , Liao, X. , Yang, X. , Luo, Y. , Lin, P. , Zeng, Q. et al. (2021) Lysine crotonylation of DgTIL1 at K72 modulates cold tolerance by enhancing DgnsLTP stability in chrysanthemum. Plant Biotechnology Journal, 19, 1125–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, H.S. , Adhikari, P.B. , Jo, H.J. , Han, J.Y. & Choi, Y.E. (2020) Enhanced monoterpene emission in transgenic orange mint (Mentha × piperita f. citrata) overexpressing a tobacco lipid transfer protein (NtLTP1). Planta, 252, 44. [DOI] [PubMed] [Google Scholar]
- Isaacs, M. , Carella, P. , Faubert, J. , Rose, J.K. & Cameron, R.K. (2016) Orthology analysis and in vivo complementation studies to elucidate the role of DIR1 during systemic acquired resistance in Arabidopsis thaliana and Cucumis sativus . Frontiers in Plant Science, 7, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq, A. , Pernot, C. , Martinez, Y. , Domergue, F. , Payre, B. , Jamet, E. et al. (2017) The Arabidopsis lipid transfer protein 2 (AtLTP2) Is involved in cuticle‐cell wall interface integrity and in etiolated hypocotyl permeability. Frontiers in Plant Science, 8, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Z. , Gou, J. , Sun, Y. , Yuan, L. , Tang, Q. , Yang, X. et al. (2010) Enhanced resistance to fungal pathogens in transgenic Populus tomentosa Carr. by overexpression of an nsLTP‐like antimicrobial protein gene from motherwort (Leonurus japonicus). Tree Physiology, 30, 1599–1605. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Wang, W. , Xie, Q. , Liu, N. , Liu, L. , Wang, D. et al. (2017) Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science, 356, 1172–1175. [DOI] [PubMed] [Google Scholar]
- Jing, R. , Wen, T. , Liao, C. , Xue, L. , Liu, F. , Yu, L. et al. (2021) DeepT3 2.0: improving type III secreted effector predictions by an integrative deep learning framework. NAR Genomics and Bioinformatics, 3, b86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, H.W. , Tschaplinski, T.J. , Wang, L. , Glazebrook, J. & Greenberg, J.T. (2009) Priming in systemic plant immunity. Science, 324, 89–91. [DOI] [PubMed] [Google Scholar]
- Kader, J.C. (1975) Proteins and the intracellular exchange of lipids: stimulation of phospholipid exchange between mitochondria and microssomal fractions by proteins isolated from potato tuber. BiochimicaI et Biophysica Acta Biomembranes, 380, 31–44. [PubMed] [Google Scholar]
- Kalla, R. , Shimamoto, K. , Potter, R. , Nielsen, P.S. , Linnestad, C. & Olsen, O.A. (1994) The promoter of the barley aleurone‐specific gene encoding a putative 7 kDa lipid transfer protein confers aleurone cell‐specific expression in transgenic rice. The Plant Journal, 6, 849–860. [DOI] [PubMed] [Google Scholar]
- Kielbowicz‐Matuk, A. , Rey, P. & Rorat, T. (2008) The organ‐dependent abundance of a Solanum lipid transfer protein is up‐regulated upon osmotic constraints and associated with cold acclimation ability. Journal of Experimental Botany, 59, 2191–2203. [DOI] [PubMed] [Google Scholar]
- Kim, H. , Lee, S.B. , Kim, H.J. , Min, M.K. , Hwang, I. & Suh, M.C. (2012) Characterization of glycosylphosphatidylinositol‐anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana . Plant & Cell Physiology, 53, 1391–1403. [DOI] [PubMed] [Google Scholar]
- Kim, T.H. , Park, J.H. , Kim, M.C. & Cho, S.H. (2008) Cutin monomer induces expression of the rice OsLTP5 lipid transfer protein gene. Journal of Plant Physiology, 165, 345–349. [DOI] [PubMed] [Google Scholar]
- Kirubakaran, S.I. , Begum, S.M. , Ulaganathan, K. & Sakthivel, N. (2008) Characterization of a new antifungal lipid transfer protein from wheat. Plant Physiology and Biochemistry, 46, 918–927. [DOI] [PubMed] [Google Scholar]
- Kouidri, A. , Baumann, U. , Okada, T. , Baes, M. , Tucker, E.J. & Whitford, R. (2018) Wheat TaMs1 is a glycosylphosphatidylinositol‐anchored lipid transfer protein necessary for pollen development. BMC Plant Biology, 18, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk, N. , Smith, J. , Bazanova, N. , Pyvovarenko, T. , Singh, R. , Shirley, N. et al. (2012) Characterization of the wheat gene encoding a grain‐specific lipid transfer protein TdPR61, and promoter activity in wheat, barley and rice. Journal of Experimental Botany, 63, 2025–2040. [DOI] [PubMed] [Google Scholar]
- Kovalchuk, N. , Smith, J. , Pallotta, M. , Singh, R. , Ismagul, A. , Eliby, S. et al. (2009) Characterization of the wheat endosperm transfer cell‐specific protein TaPR60. Plant Molecular Biology, 71, 81–98. [DOI] [PubMed] [Google Scholar]
- Lascombe, M.B. , Bakan, B. , Buhot, N. , Marion, D. , Blein, J.P. , Larue, V. et al. (2008) The structure of "defective in induced resistance" protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Science, 17, 1522–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasica, A.M. , Ksiazek, M. , Madej, M. & Potempa, J. (2017) The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Frontiers in Cellular and Infection Microbiology, 7, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y. , Min, K. , Cha, H. , Shin, D.H. , Hwang, K.Y. & Suh, S.W. (1998) Rice non‐specific lipid transfer protein: the 1.6 A crystal structure in the unliganded state reveals a small hydrophobic cavity. Journal of Molecular Biology, 276, 437–448. [DOI] [PubMed] [Google Scholar]
- Lee, S.B. & Suh, M.C. (2018) Disruption of glycosylphosphatidylinositol‐anchored lipid transfer protein 15 affects seed coat permeability in Arabidopsis . The Plant Journal, 96, 1206–1217. [DOI] [PubMed] [Google Scholar]
- Lee, S.B. , Go, Y.S. , Bae, H.J. , Park, J.H. , Cho, S.H. , Cho, H.J. et al. (2009) Disruption of glycosylphosphatidylinositol‐anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola . Plant Physiology, 150, 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, L. , Chen, L. , Shi, X. , Li, Y. , Wang, J. , Chen, D. et al. (2014) A nodule‐specific lipid transfer protein AsE246 participates in transport of plant‐synthesized lipids to symbiosome membrane and is essential for nodule organogenesis in Chinese milk vetch. Plant Physiology, 164, 1045–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Lopato, S. , Hrmova, M. , Pickering, M. , Shirley, N. , Koltunow, A.M. et al. (2014) Expression patterns and protein structure of a lipid transfer protein END1 from Arabidopsis . Planta, 240, 1319–1334. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Ai, G. , Shen, D. , Zou, F. , Wang, J. , Bai, T. et al. (2019) A Phytophthora capsici effector targets ACD11 binding partners that regulate ROS‐mediated defense response in Arabidopsis . Molecular Plant, 12, 565–581. [DOI] [PubMed] [Google Scholar]
- Li, R. , Xia, J. , Xu, Y. , Zhao, X. , Liu, Y.G. & Chen, Y. (2014) Characterization and genetic mapping of a Photoperiod‐sensitive dwarf 1 locus in rice (Oryza sativa L.). Theoretical and Applied Genetics, 127, 241–250. [DOI] [PubMed] [Google Scholar]
- Lim, G.H. , Liu, H. , Yu, K. , Liu, R. , Shine, M.B. , Fernandez, J. et al. (2020) The plant cuticle regulates apoplastic transport of salicylic acid during systemic acquired resistance. Science Advances, 6, z478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, G.H. , Shine, M.B. , de Lorenzo, L. , Yu, K. , Cui, W. , Navarre, D. et al. (2016) Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host & Microbe, 19, 541–549. [DOI] [PubMed] [Google Scholar]
- Lin, K.F. , Liu, Y.N. , Hsu, S.T. , Samuel, D. , Cheng, C.S. , Bonvin, A.M. et al. (2005) Characterization and structural analyses of nonspecific lipid transfer protein 1 from mung bean. Biochemistry, 44, 5703–5712. [DOI] [PubMed] [Google Scholar]
- Liu, F. , Xiong, X. , Wu, L. , Fu, D. , Hayward, A. , Zeng, X. et al. (2014) BraLTP1, a lipid transfer protein gene involved in epicuticular wax deposition, cell proliferation and flower development in Brassica napus . PLoS One, 9, e110272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Ravichandran, S. , Teh, O.K. , McVey, S. , Lilley, C. , Teresinski, H.J. et al. (2017) The RING‐type E3 ligase XBAT35.2 is involved in cell death induction and pathogen response. Plant Physiology, 175, 1469–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K.H. & Lin, T.Y. (2003) Cloning and characterization of two novel lipid transfer protein I genes in Vigna radiata . DNA Sequence, 14, 420–426. [DOI] [PubMed] [Google Scholar]
- Luginbuehl, L.H. , Menard, G.N. , Kurup, S. , Van Erp, H. , Radhakrishnan, G.V. , Breakspear, A. et al. (2017) Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science, 356, 1175–1178. [DOI] [PubMed] [Google Scholar]
- Madni, Z.K. , Tripathi, S.K. & Salunke, D.M. (2020) Structural insights into the lipid transfer mechanism of a non‐specific lipid transfer protein. The Plant Journal, 102, 340–352. [DOI] [PubMed] [Google Scholar]
- Maeda, L. , Anand, K. , Chiapparino, A. , Kumar, A. , Poletto, M. , Kaksonen, M. et al. (2013) Interactome map uncovers phosphatidylserine transport by oxysterol‐binding proteins. Nature, 501, 257–261. [DOI] [PubMed] [Google Scholar]
- Maldonado, A.M. , Doerner, P. , Dixon, R.A. , Lamb, C.J. & Cameron, R.K. (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis . Nature, 419, 399–403. [DOI] [PubMed] [Google Scholar]
- Malinovsky, F.G. , Brodersen, P. , Fiil, B.K. , McKinney, L.V. , Thorgrimsen, S. , Beck, M. et al. (2010) Lazarus1, a DUF300 protein, contributes to programmed cell death associated with Arabidopsis acd11 and the hypersensitive response. PLoS One, 5, e12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin, J.E. , Bin‐Umer, M.A. , Widiez, T. , Finn, D. , McCormick, S. & Tumer, N.E. (2015) A lipid transfer protein increases the glutathione content and enhances Arabidopsis resistance to a trichothecene mycotoxin. PLoS One, 10, e130204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin, J.E. , Darwish, N.I. , Garcia‐Sanchez, J. , Tyagi, N. , Trick, H.N. , McCormick, S. et al. (2021) A Lipid transfer protein has antifungal and antioxidant activity and suppresses Fusarium head blight disease and DON accumulation in transgenic wheat. Phytopathology, 111, 671–683. [DOI] [PubMed] [Google Scholar]
- Missaoui, K. , Gonzalez‐Klein, Z. , Pazos‐Castro, D. , Hernandez‐Ramirez, G. , Garrido‐Arandia, M. , Brini, F. et al. (2022) Plant non‐specific lipid transfer proteins: an overview. Plant Physiology and Biochemistry, 171, 115–127. [DOI] [PubMed] [Google Scholar]
- Munch, D. , Teh, O.K. , Malinovsky, F.G. , Liu, Q. , Vetukuri, R.R. , El, K.F. et al. (2015) Retromer contributes to immunity‐associated cell death in Arabidopsis . The Plant Cell, 27, 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrot, R. , Jozefiak, D. , Sip, A. , Kuzma, D. , Musidlak, O. & Gozdzicka‐Jozefiak, A. (2017) Isolation and characterization of a non‐specific lipid transfer protein from Chelidonium majus L. latex. International Journal of Biological Macromolecules, 104, 554–563. [DOI] [PubMed] [Google Scholar]
- Nazeer, M. , Waheed, H. , Saeed, M. , Ali, S.Y. , Choudhary, M.I. , Ul‐Haq, Z. et al. (2019) Purification and characterization of a nonspecific lipid transfer protein 1 (nsLTP1) from ajwain (Trachyspermum ammi) seeds. Scientific Reports, 9, 4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland, J. , Feron, R. , Huisman, B.A. , Fasolino, A. , Hilbers, C.W. , Derksen, J. et al. (2005) Lipid transfer proteins enhance cell wall extension in tobacco. The Plant Cell, 17, 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat, L.A. , Oyarburo, N. , Cimmino, C. , Pinedo, M.L. & de la Canal, L. (2015) On the role of a Lipid‐Transfer Protein. Arabidopsis ltp3 mutant is compromised in germination and seedling growth. Plant Signaling & Behavior, 10, e1105417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat, L. , Burbach, C. , Baluska, F. & de la Canal, L. (2012) An extracellular lipid transfer protein is relocalized intracellularly during seed germination. Journal of Experimental Botany, 63, 6555–6563. [DOI] [PubMed] [Google Scholar]
- Pallotta, M.A. , Warner, P. , Kouidri, A. , Tucker, E.J. , Baes, M. , Suchecki, R. et al. (2019) Wheat ms5 male‐sterility is induced by recessive homoeologous A and D genome non‐specific lipid transfer proteins. The Plant Journal, 99, 673–685. [DOI] [PubMed] [Google Scholar]
- Palma, K. , Thorgrimsen, S. , Malinovsky, F.G. , Fiil, B.K. , Nielsen, H.B. , Brodersen, P. et al. (2010) Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathogens, 6, e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y. , Li, J. , Jiao, L. , Li, C. , Zhu, D. & Yu, J. (2016) A non‐specific Setaria italica lipid transfer protein gene plays a critical role under abiotic stress. Frontiers in Plant Science, 7, 1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorczyk‐Szlenkier, M. & Bednarek, P. (2021) UGT76B1 controls the growth‐immunity trade‐off during systemic acquired resistance. Molecular Plant, 14, 544–546. [DOI] [PubMed] [Google Scholar]
- Petersen, N.H. , Joensen, J. , McKinney, L.V. , Brodersen, P. , Petersen, M. , Hofius, D. et al. (2009) Identification of proteins interacting with Arabidopsis ACD11. Journal of Plant Physiology, 166, 661–666. [DOI] [PubMed] [Google Scholar]
- Pii, Y. , Astegno, A. , Peroni, E. , Zaccardelli, M. , Pandolfini, T. & Crimi, M. (2009) The Medicago truncatula N5 gene encoding a root‐specific lipid transfer protein is required for the symbiotic interaction with Sinorhizobium meliloti . Molecular Plant‐Microbe Interactions, 22, 1577–1587. [DOI] [PubMed] [Google Scholar]
- Pii, Y. , Molesini, B. , Masiero, S. & Pandolfini, T. (2012) The non‐specific lipid transfer protein N5 of Medicago truncatula is implicated in epidermal stages of Rhizobium‐host interaction. BMC Plant Biology, 12, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke, A. , Datta, S. & Persak, H. (2014) Salt stress in Arabidopsis: lipid transfer protein AZI1 and its control by mitogen‐activated protein kinase MPK3. Molecular Plant, 7, 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke, A. , Xue, H. , Persak, H. , Datta, S. & Seifert, G.J. (2016) Post‐translational modification and secretion of azelaic acid induced 1 (AZI1), a hybrid proline‐rich protein from Arabidopsis . International Journal of Molecular Sciences, 17, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponchet, M. , Panabieres, F. , Milat, M. , Mikes, V. , Montillet, J.L. , Suty, L. et al. (1999) Are elicitins cryptograms in plant–oomycete communications? Cellular and Molecular Life Sciences, 56, 1020–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocka, I. , Baldwin, T.C. & Kurczynska, E.U. (2012) Distribution of lipid transfer protein 1 (LTP1) epitopes associated with morphogenic events during somatic embryogenesis of Arabidopsis thaliana . Plant Cell Reports, 31, 2031–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyee, J. & Kolattukudy, P.E. (1995) The gene for the major cuticular wax‐associated protein and three homologous genes from broccoli (Brassica oleracea) and their expression patterns. The Plant Journal, 7, 49–59. [DOI] [PubMed] [Google Scholar]
- Rich, M.K. , Vigneron, N. , Libourel, C. , Keller, J. , Xue, L. , Hajheidari, M. et al. (2021) Lipid exchanges drove the evolution of mutualism during plant terrestrialization. Science, 372, 864–868. [DOI] [PubMed] [Google Scholar]
- Riederer, M. & Schreiber, L. (2001) Protecting against water loss: analysis of the barrier properties of plant cuticles. Journal of Experimental Botany, 52, 2023–2032. [DOI] [PubMed] [Google Scholar]
- Riedlmeier, M. , Ghirardo, A. , Wenig, M. , Knappe, C. , Koch, K. , Georgii, E. et al. (2017) Monoterpenes support systemic acquired resistance within and between plants. The Plant Cell, 29, 1440–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy‐Barman, S. , Sautter, C. & Chattoo, B.B. (2006) Expression of the lipid transfer protein Ace‐AMP1 in transgenic wheat enhances antifungal activity and defense responses. Transgenic Research, 15, 435–446. [DOI] [PubMed] [Google Scholar]
- Royo, J. , Gomez, E. , Sellam, O. , Gerentes, D. , Paul, W. & Hueros, G. (2014) Two maize END‐1 orthologs, BETL9 and BETL9like, are transcribed in a non‐overlapping spatial pattern on the outer surface of the developing endosperm. Frontiers in Plant Science, 5, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safi, H. , Saibi, W. , Alaoui, M.M. , Hmyene, A. , Masmoudi, K. , Hanin, M. et al. (2015) A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana . Plant Physiology and Biochemistry, 89, 64–75. [DOI] [PubMed] [Google Scholar]
- Salminen, T.A. , Blomqvist, K. & Edqvist, J. (2016) Lipid transfer proteins: classification, nomenclature, structure, and function. Planta, 244, 971–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel, D. , Liu, Y.J. , Cheng, C.S. & Lyu, P.C. (2002) Solution structure of plant nonspecific lipid transfer protein‐2 from rice (Oryza sativa). Journal of Biological Chemistry, 277, 35267–35273. [DOI] [PubMed] [Google Scholar]
- Sarowar, S. , Kim, Y.J. , Kim, K.D. , Hwang, B.K. , Ok, S.H. & Shin, J.S. (2009) Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long‐distance systemic signaling in tobacco. Plant Cell Reports, 28, 419–427. [DOI] [PubMed] [Google Scholar]
- Sawano, Y. , Hatano, K. , Miyakawa, T. , Komagata, H. , Miyauchi, Y. , Yamazaki, H. et al. (2008) Proteinase inhibitor from ginkgo seeds is a member of the plant nonspecific lipid transfer protein gene family. Plant Physiology, 146, 1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, A.J. , Sathoff, A.E. , Holl, C. , Bauer, B. , Samac, D.A. & Carter, C.J. (2018) The major nectar protein of Brassica rapa is a non‐specific lipid transfer protein, BrLTP2.1, with strong antifungal activity. Journal of Experimental Botany, 69, 5587–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnake, A. , Hartmann, M. , Schreiber, S. , Malik, J. , Brahmann, L. , Yildiz, I. et al. (2020) Inducible biosynthesis and immune function of the systemic acquired resistance inducer N‐hydroxypipecolic acid in monocotyledonous and dicotyledonous plants. Journal of Experimental Botany, 71, 6444–6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sels, J. , Mathys, J. , De Coninck, B.M. , Cammue, B.P. & De Bolle, M.F. (2008) Plant pathogenesis‐related (PR) proteins: a focus on PR peptides. Plant Physiology and Biochemistry, 46, 941–950. [DOI] [PubMed] [Google Scholar]
- Simanshu, D.K. , Zhai, X. , Munch, D. , Hofius, D. , Markham, J.E. , Bielawski, J. et al. (2014) Arabidopsis accelerated cell death 11, ACD11, is a ceramide‐1‐phosphate transfer protein and intermediary regulator of phytoceramide levels. Cell Reports, 6, 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, A.A. , Costa, A.S. , Campos, D. , Batista, A. , Sales, G. , Nogueira, N. et al. (2018) Lipid transfer protein isolated from noni seeds displays antibacterial activity in vitro and improves survival in lethal sepsis induced by CLP in mice. Biochimie, 149, 9–17. [DOI] [PubMed] [Google Scholar]
- Sun, J.Y. , Gaudet, D.A. , Lu, Z.X. , Frick, M. , Puchalski, B. & Laroche, A. (2008) Characterization and antifungal properties of wheat nonspecific lipid transfer proteins. Molecular Plant‐Microbe Interactions, 21, 346–360. [DOI] [PubMed] [Google Scholar]
- Tao, Y. , Zou, T. , Zhang, X. , Liu, R. , Chen, H. , Yuan, G. et al. (2021) Secretory lipid transfer protein OsLTPL94 acts as a target of EAT1 and is required for rice pollen wall development. The Plant Journal, 108, 358–377. [DOI] [PubMed] [Google Scholar]
- Tapia, G. , Morales‐Quintana, L. , Parra, C. , Berbel, A. & Alcorta, M. (2013) Study of nsLTPs in Lotus japonicus genome reveal a specific epidermal cell member (LjLTP10) regulated by drought stress in aerial organs with a putative role in cutin formation. Plant Molecular Biology, 82, 485–501. [DOI] [PubMed] [Google Scholar]
- Tassin, S. , Broekaert, W.F. , Marion, D. , Acland, D.P. , Ptak, M. , Vovelle, F. et al. (1998) Solution structure of Ace‐AMP1, a potent antimicrobial protein extracted from onion seeds. Structural analogies with plant nonspecific lipid transfer proteins. Biochemistry, 37, 3623–3637. [DOI] [PubMed] [Google Scholar]
- Tian, N. , Liu, F. , Wang, P. , Yan, X. , Gao, H. , Zeng, X. et al. (2018) Overexpression of BraLTP2, a lipid transfer protein of Brassica napus, results in increased trichome density and altered concentration of secondary metabolites. International Journal of Molecular Sciences, 19, 1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen, M.A. , Verhees, J.A. , Schmidt, E.D. , van Kammen, A. & de Vries, S.C. (1997) AtLTP1 luciferase expression during carrot somatic embryogenesis. The Plant Journal, 12, 1213–1221. [DOI] [PubMed] [Google Scholar]
- Tucker, E.J. , Baumann, U. , Kouidri, A. , Suchecki, R. , Baes, M. , Garcia, M. et al. (2017) Molecular identification of the wheat male fertility gene Ms1 and its prospects for hybrid breeding. Nature Communications, 8, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Gao, H. , Chu, Z. , Ji, C. , Xu, Y. , Cao, W. et al. (2021) A nonspecific lipid transfer protein, StLTP10, mediates resistance to Phytophthora infestans in potato. Molecular Plant Pathology, 22, 48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Zang, X.S. , Kabir, M.R. , Liu, K.L. , Liu, Z.S. , Ni, Z.F. et al. (2014) A wheat lipid transfer protein 3 could enhance the basal thermotolerance and oxidative stress resistance of Arabidopsis . Gene, 550, 18–26. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Sun, Y. , Chang, J. , Zheng, F. , Pei, H. , Yi, Y. et al. (2016) Regulatory function of Arabidopsis lipid transfer protein 1 (LTP1) in ethylene response and signaling. Plant Molecular Biology, 91, 471–484. [DOI] [PubMed] [Google Scholar]
- Wang, S.Y. , Wu, J.H. , Ng, T.B. , Ye, X.Y. & Rao, P.F. (2004) A non‐specific lipid transfer protein with antifungal and antibacterial activities from the mung bean. Peptides, 25, 1235–1242. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Zhou, W. , Lu, Z. , Ouyang, Y. & Yao, J. (2015) A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Science, 239, 200–208. [DOI] [PubMed] [Google Scholar]
- Xia, Y. , Gao, Q.M. , Yu, K. , Lapchyk, L. , Navarre, D. , Hildebrand, D. et al. (2009) An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host & Microbe, 5, 151–165. [DOI] [PubMed] [Google Scholar]
- Xia, Y. , Yu, K. , Gao, Q.M. , Wilson, E.V. , Navarre, D. , Kachroo, P. et al. (2012) Acyl coA binding proteins are required for cuticle formation and plant responses to microbes. Frontiers in Plant Science, 3, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y. , Yu, K. , Navarre, D. , Seebold, K. , Kachroo, A. & Kachroo, P. (2010) The glabra1 mutation affects cuticle formation and plant responses to microbes. Plant Physiology, 154, 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Q. , Lott, A.A. , Assmann, S.M. & Chen, S. (2021) Advances and perspectives in the metabolomics of stomatal movement and the disease triangle. Plant Science, 302, 110697. [DOI] [PubMed] [Google Scholar]
- Xu, Y. , Zheng, X. , Song, Y. , Zhu, L. , Yu, Z. , Gan, L. et al. (2018) NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum . Scientific Reports, 8, 8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Li, J. , Li, X. , She, R. & Pei, Y. (2006) Isolation and characterization of a novel thermostable non‐specific lipid transfer protein‐like antimicrobial protein from motherwort (Leonurus japonicus Houtt) seeds. Peptides, 27, 3122–3128. [DOI] [PubMed] [Google Scholar]
- Yu, K. , Soares, J.M. , Mandal, M.K. , Wang, C. , Chanda, B. , Gifford, A.N. et al. (2013) A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic‐acid‐induced systemic immunity. Cell Reports, 3, 1266–1278. [DOI] [PubMed] [Google Scholar]
- Zaidi, M.A. , O'Leary, S. , Gagnon, C. , Chabot, D. , Wu, S. , Hubbard, K. et al. (2020) A triticale tapetal non‐specific lipid transfer protein (nsLTP) is translocated to the pollen cell wall. Plant Cell Reports, 39, 1185–1197. [DOI] [PubMed] [Google Scholar]
- Zhai, X. , Gao, Y.G. , Mishra, S.K. , Simanshu, D.K. , Boldyrev, I.A. , Benson, L.M. et al. (2017) Phosphatidylserine stimulates ceramide 1‐phosphate (C1P) intermembrane transfer by C1P transfer proteins. Journal of Biological Chemistry, 292, 2531–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Liang, W. , Yin, C. , Zong, J. , Gu, F. & Zhang, D. (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiology, 154, 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Bi, W. , Zhao, S. , Su, J. , Li, M. , Ma, L. et al. (2021) Wheat apoplast‐localized lipid transfer protein TaLTP3 enhances defense responses against Puccinia triticina . Frontiers in Plant Science, 12, 771806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Wang, S. , Qin, J. , Sun, C. & Liu, F. (2020) The lipid transfer protein OsLTPL159 is involved in cold tolerance at the early seedling stage in rice. Plant Biotechnology Journal, 18, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D. , Li, Y. , Wang, X. , Xie, F. , Chen, D. , Ma, B. et al. (2019) Mesorhizobium huakuii HtpG interaction with nsLTP AsE246 is required for symbiotic nitrogen fixation. Plant Physiology, 180, 509–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q. , Meng, Q. , Tan, X. , Ding, W. , Ma, K. , Xu, Z. et al. (2021) Protein phosphorylation changes during systemic acquired resistance in Arabidopsis thaliana . Frontiers in Plant Science, 12, 748287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Li, Z. , Xu, H. , Zhou, M. , Du, L. & Zhang, Z. (2012) Overexpression of wheat lipid transfer protein gene TaLTP5 increases resistances to Cochliobolus sativus and Fusarium graminearum in transgenic wheat. Functional & Integrative Genomics, 12, 481–488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


