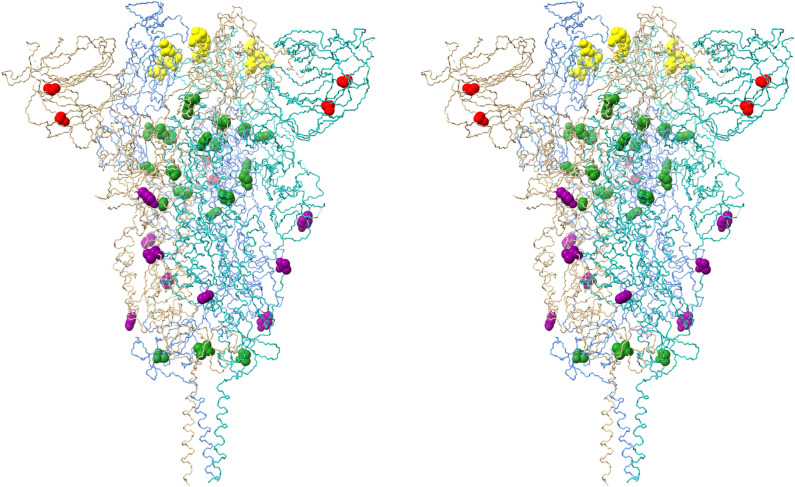
Figure 6.
Stereo pair view of spike protein receptor trimer illustrating the locations of all Omicron spike protein mutations (excepting RBD contact surface mutations present in Fig. 5). Monomer A is blue, monomer B is salmon and monomer C is aquamarine. All mutated residues are rendered as CPK models. Yellow residues (S371L, S373P, S375F, K417N) are located in the RBD where they may alter domain packing and/or monomer–monomer interactions. Green residues (T547K, N764K, N856K, Q954H, N969K, L981F, I1081V) are located at or near monomer–monomer interfaces in the receptor trimer assembly. Red residues (A67V, T95I) are located in the interior of the N-terminal domain. Purple residues (N655Y, A701V, D796Y) are located on the exterior surface of the trimer assembly in regions that are potential antibody binding sites. The homology model is based on reference spike protein trimer structure PDB ID: 7KRS.