Figure 1.
Overview of SigProfilerExtractor
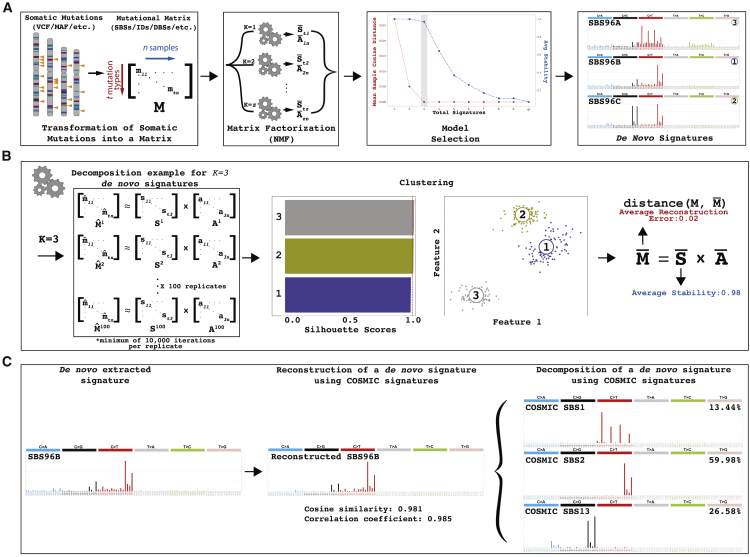
(A) SigProfilerExtractor’s general workflow is outlined starting from an input of somatic mutations and resulting in an output of de novo mutational signatures. An example is shown for a solution with three de novo signatures. Somatic mutations are first converted into a mutational matrix M. Subsequently, the matrix is factorized with different ranks using nonnegative matrix factorization. Model selection is applied to identify the optimal factorization rank based on each solution’s stability and its reconstruction of the original data.
(B) Schematic representation for an example decomposition with a factorization rank of k = 3 reflecting three operative mutational signatures. By default, SigProfilerExtractor performs 100 independent nonnegative matrix factorizations with the matrix M being Poisson resampled and normalized (denoted by “ˆ”) prior to each factorization. Partition clustering of the 100 factorizations is used to evaluate the factorization stability rank, measured in silhouette values; clustering can also be presented as two-dimensional projections revealing more similar mutational signatures as shown for the three example signatures. The centroid of the clustered solutions (denoted by “–”) is compared with the original matrix M.
(C) All identified de novo signatures are matched to a combination of known COSMIC mutational signatures. An example is given for de novo extracted signature SBS96B, which matches a combination of COSMIC signatures SBS1, SBS2, and SBS13.