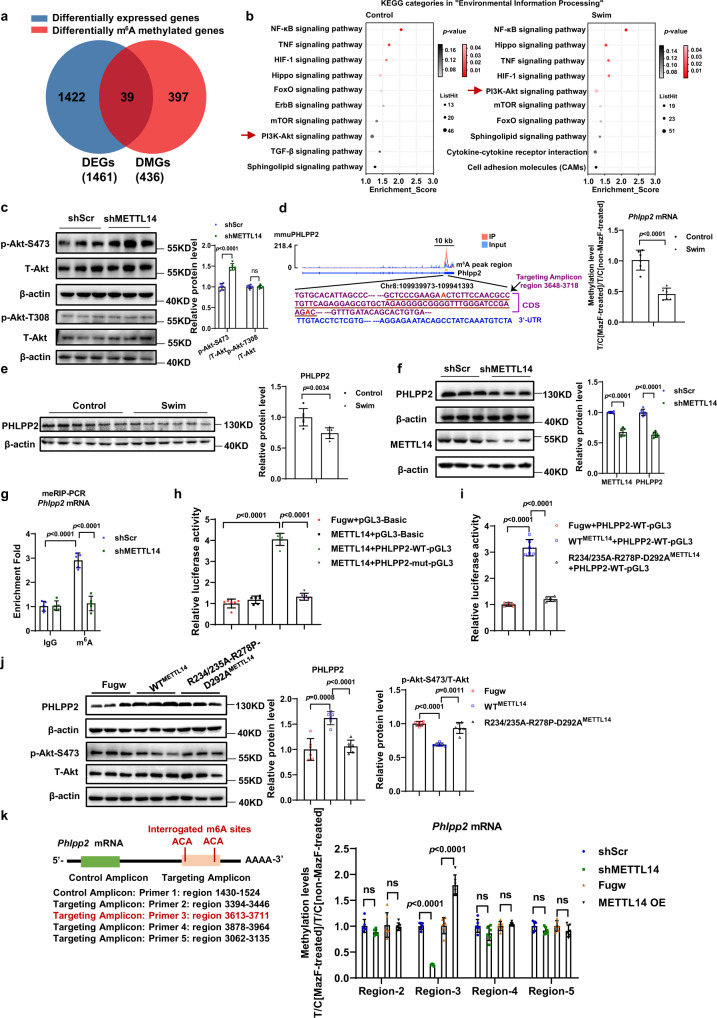
Fig. 7. METTL14 knockdown inhibits Phlpp2 mRNA m6A modifications and activates Akt-S473.
a Venn diagram showing the comparison of differentially methylated transcripts and differentially expressed transcripts identified in the exercise-induced cardiac hypertrophy murine model. DMG, differentially m6A methylated genes. DEG, differentially expressed genes. b KEGG analysis of significantly enriched m6A methylated peaks (p < 0.05; fold-change > 2.0) in control group and swim group, respectively. c Representative western blot and statistical data of phosphorylation levels of Akt-S473, Akt-T308 in NRCMs with or without METTL14 knockdown (n = 6 wells/group). d The methylation levels of Phlpp2 mRNA in swim and control mouse hearts measured by MazF-qPCR (n = 6 mice/group). The levels of a targeted amplicon (labeled “T”) is measured against a control (labeled “C”) amplicon in a MazF-digested sample and normalized against a nondigested sample. e Western blot analyses of PHLPP2 in whole lysates isolated from swim and control murine hearts (n = 6 mice/group). f Western blot analyses of PHLPP2 and METTL14 in NRCM with or without METTL14 inhibition (n = 6 wells/group). g m6A enrichment levels in Phlpp2 mRNA in H9C2 cardiomyocytes treated as indicated using m6A-RIP (meRIP)-qPCR (n = 5/group). h Relative luciferase activity of Rattus PHLPP2 wild-type (PHLPP2-WT-pGL3) or m6A mutant (PHLPP2-mut-pGL3) reporter gene with or without Rattus METTL14 overexpression (n = 6/group). i Relative luciferase activity of Rattus PHLPP2-WT-pGL3 reporter gene with Rattus wild-type METTL14 (WTMETTL14) or mutant METTL14 (R234/235A-R278P-D292AMETTL14) overexpression (n = 6/group). j Representative western blot and statistical data of PHLPP2 and phosphorylation level of Akt-S473 in NRCMs with WTMETTL14 or R234/235A-R278P-D292AMETTL14 overexpression (n = 6/group). k MazF-qPCR identified the methylation modification region of Phlpp2 mRNA which is specifically regulated by METTL14 (n = 6/group). The levels of a targeted amplicon (labeled “T”) is measured against a control (labeled “C”) amplicon in a MazF-digested sample and normalized against a nondigested sample. NRCM, neonatal rat cardiomyocyte; shScr, Scramble short hairpin RNA; shMETTL14, METTL14 short hairpin RNA to knockdown METTL14. ns, nonstatistically significant. All data are expressed as means ± SD. c–f, k Independent-sample t-test, two-sided; g two-way ANOVA followed by Tukey’s post hoc test; h, j one-way ANOVA followed by Bonferroni test; i one-way ANOVA followed by Dunnett T3 test.) Source data are provided as a Source Data file.