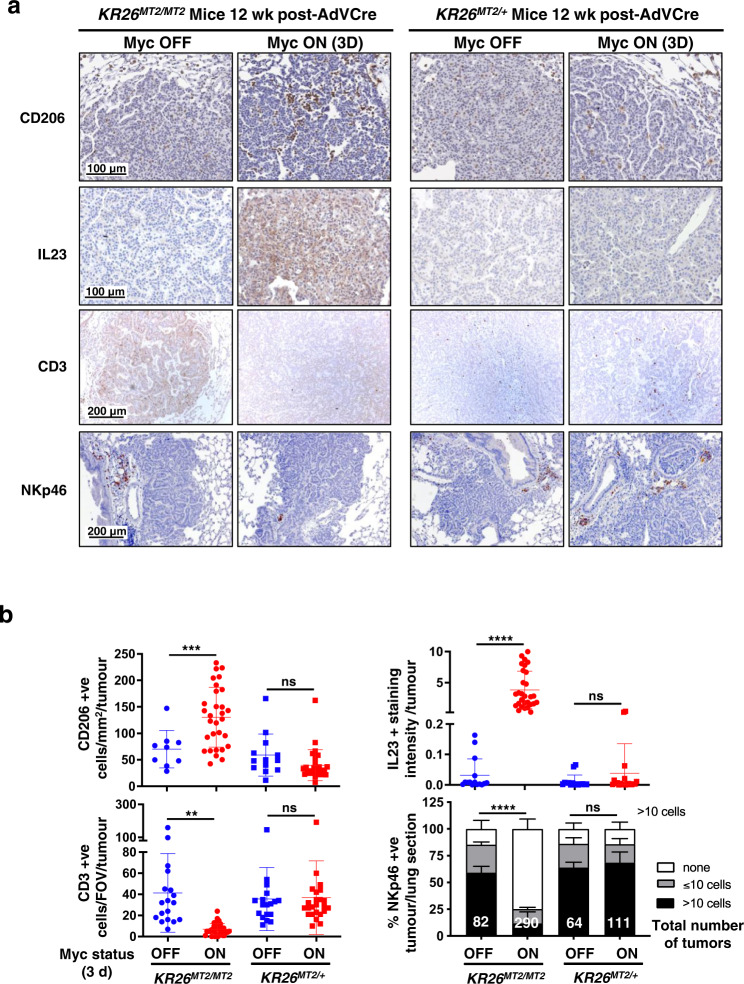
Fig. 4. A minimum threshold level of Myc is required to engage instructive stromal signals that drive transition from indolent pre-tumour to invasive neoplasia.
a Representative IHC analysis of in sections of lungs harvested from KR26MT2/MT2 and KR26MT2/+ mice 12 weeks after activation of KRasG12D, either without or with activation of MycERT2 for 3 days. IHC staining is shown for CD206+ macrophages (top row), IL23 (second row), CD3+ T cells (third row), and NKp46+ NK cells (bottom row) in sections of lungs harvested from KR26MT2/MT2 and KR26MT2/+ mice 12 weeks after activation of KrasG12D, either without or with activation of MycERT2 for 3 days. b Quantification of CD206, IL23, CD3 and NKp46 immunohistochemical staining (IHC) in sections of lungs described in A. Results depict mean ± SD of independent tumours from 3-6 mice per treatment group. IL23 + staining intensity was normalised to the number of nuclei per FOV (Field of view) as described in ref. 66. For NKp46 staining, tumours connected to clearly distinguishable vascular and airway regions were considered; the number of tumour-associated NKp46+ cells per tumour per lung section were counted. Data were analysed using unpaired t-test with Welch’s correction and two-tailed analysis (CD206, IL23, CD3) or two-way ANOVA (NKp46). For CD206 staining, n = 9 independent tumours (3 mice) for Myc OFF and n = 30 independent tumours (6 mice) for Myc ON KR26MT2/MT2 groups with p = 0.0009; n = 13 independent tumours (3 mice) for Myc OFF and n = 27 independent tumours (4 mice) for Myc ON KR26MT2/+ groups with p = 0.1427. For IL23 staining, n = 15 independent tumours (3 mice) for Myc OFF and n = 29 independent tumours (6 mice) for Myc ON KR26MT2/MT2 groups with p < 0.0001; n = 15 independent tumours (3 mice) for Myc OFF and n = 20 independent tumours (4 mice) for Myc ON KR26MT2/+ groups with p = 0.2339. For CD3 staining, n = 18 independent tumours (3 mice) for Myc OFF and n = 32 independent tumours (6 mice) for Myc ON KR26MT2/MT2 groups with p = 0.0011; n = 18 independent tumours (3 mice) for Myc OFF and n = 24 independent tumours (4 mice) for Myc ON KR26MT2/+ groups with p = 0.8982. For NKp46 staining, n = 82 independent tumours (3 mice) for Myc OFF and n = 290 independent tumours (6 mice) for Myc ON KR26MT2/MT2 groups with adjusted p < 0.0001 for > 10 cells; n = 64 independent tumours (3 mice) for Myc OFF and n = 111 independent tumours (4 mice) for Myc ON KR26MT2/+ groups with adjusted p = 0.8311 for >10 cells. **p < 0.01, ***p < 0.001, ****p < 0.0001, ns non-significant, SD standard deviation. Source data are provided as a Source Data file.