Table 1.
Free binding energies of the top 20 scoring hits and their plant source along with the co-crystalized ligand, leflunomide and the reference drug, brequinar, obtained through molecular docking with hDHODH crystal structure (3G0U), in addition to, types of binding interactions between ligands and critical amino acid residues in the hDHODH binding site.
| S. no | Name of top scored compounds (its plant source) | Binding free energy (Kcal mol-1) | Type of bindig interactions | Amino acid residues involved in protein ligand interaction |
|---|---|---|---|---|
| 1 |
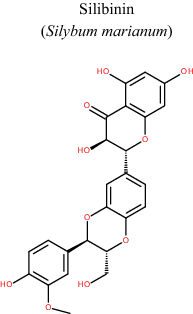
|
− 14.857 | Hydrogen bonding (side chain) | TYR356, ARG136 |
| Polar interaction | GLN47, HIE56, THR360 | |||
| Hydrophobic interaction | LEU46, MET43, LEU42, PHE62, LEU58, PRO364, TYR38, LEU68, ALA59, LEU359, ALA55, PHE98, TYR356, TYR147, VAL143, VAL134, PRO52 | |||
| Pi-Pi stacking interaction | PHE62 | |||
| Glycine interaction | GLY363 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| 2 |
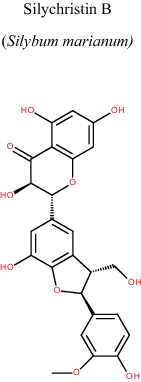
|
− 14.642 | Hydrogen bonding (backbone and side chain) | LEU359, TYR356, ARG136 |
| Charged (positive) ionic interaction | ARG136 | |||
| Pi-Pi stacking interaction | TYR38 | |||
| Glycine interaction | GLY363 | |||
| Hydrophobic interaction | LEU46, MET43, LEU359, PRO364, TYR38, LEU42, PRO69, LEU68, PHE62, LEU58, ALA59, ALA55, PHE98, TYR356, TYR147, VAL143, VAL134, PRO52 | |||
| Polar interaction | GLN47, HIE56, THR360 | |||
| 3 |
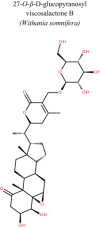
|
− 14.293 | Hydrogen bonding (side chain) | ARG136 |
| Hydrophobic interaction | LEU46, MET43, ALA59, LEU58, LEU42, PHE62, TYR38, PRO364, LEU68, PHE98, LEU359, ALA55, PRO52, VAL143, VAL134, TYR356, TYR147 | |||
| Charged (negative) ionic interaction | GLU53 | |||
| Polar interaction | GLN47, HIE56, THR360 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Glycine interaction | GLY363 | |||
| 4 |
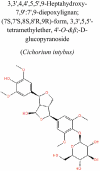
|
− 14.271 | Hydrogen bonding (side chain) | ARG136 |
| Polar interaction | THR63, HIE56, THR360, GLN47 | |||
| Pi-Pi stacking interaction | PHE62 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | LEU359, PHE62, TYR38, LEU42, MET43, LEU46, ALA55, PRO52, VAL143, VAL134, TYR147, PHE98, TYR356, ALA58, LEU68, MET111, LEU58, PRO364, PRO69 | |||
| Charged (negative) ionic interaction | GLU53 | |||
| Glycine interaction | GLY363 | |||
| 5 |
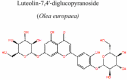
|
− 14.267 | Hydrogen bonding (side chain and backbone) | LEU42, THR360, ARG136, PRO52 |
| Polar interaction | THR45, THR360, GLN47, HIE56 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | PRO69, LEU68, TYR38, PHE62, LEU359, PRO361, ALA59, PHE98, ALA55, TYR356, VAL134, PRO52, LEU42, MET43, LEU46, LEU49,PHE37 | |||
| Glycine interaction | GLY363 | |||
| 6 |
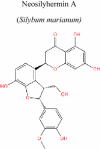
|
− 13.951 | Hydrogen bonding (side chain) | TYR356 |
| Polar interaction | THR63, HIE56, THR360, GLN47 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | TYR38, PHE62, MET111, PRO364, LEU68, LEU108, PHE98, LEU359, VAL134, TYR356, ALA55, PRO52, LEU46, MET43, ALA59, LEU58, LEU42 | |||
| Glycine interaction | GLY363 | |||
| 7 |
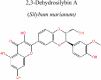
|
− 13.784 | Hydrogen bonding (side chain and backbone) | TYR356, ARG136, THR360 |
| Polar interaction | HIE56, GLN47, THR360 | |||
| Pi-Pi stacking interaction | PHE62 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | LEU58, PRO364, MET111, LEU68, ALA59, LEU359, ALA55, PHE98, TYR356, VAL134, VAL143, PRO52, LEU46, MET43, PHE62, TYR38, LEU42 | |||
| 8 |
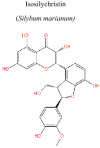
|
− 13.774 | Hydrogen bonding (side chain and backbone) | TYR356, ARG136, ALA55 |
| Polar interaction | THR63, HIE56, THR360, GLN47 | |||
| Pi-Pi stacking interaction | PHE62 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | PHE62, LEU68, ALA59, MET111, LEU359, PRO364, PHE98, VAL134, TYR356, PRO52, LEU46, MET43, TYR38, LEU42, ALA55, LEU58 | |||
| Glycine interaction | GLY363 | |||
| 9 |
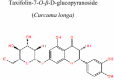
|
− 13.593 | Hydrogen bonding (side chain and backbone) | ARG136, GLN47, PRO52 |
| Polar interaction | GLN47, HIE56, THR360, THR63 | |||
| Pi-Pi stacking interaction | PHE62 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | LEU46, LEU42, MET43, PRO52, VAL134, ALA55, TYR356, PHE98, LEU359, ALA59, MET111, PRO364, LEU68, TYR38, PHE62, LEU58 | |||
| Glycine interaction | GLY363 | |||
| 10 |
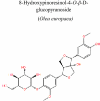
|
− 13.493 | Hydrogen bonding (side chain and backbone) | GLN47, ARG136, PRO52, TYR356 |
| Polar interaction | GLN47, THR360, HIE56 | |||
| Charged (negative) ionic interaction | GLU53 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | LEU68, PHE62, LEU58, PRO364, ALA59, PHE98, ALA55, LEU359, PRO52, VAL134, VAL143, TYR356, LEU46, MET43, TYR38, LEU42 | |||
| Glycine interaction | GLY363 | |||
| 11 |
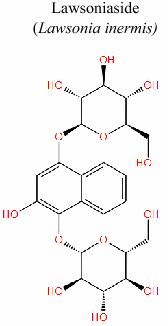
|
− 13.484 | Hydrogen bonding (side chain and backbone) | TYR38, THR360, ARG136, PRO52 |
| Polar interaction | THR63, THR360, GLN47, HIE56 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | PRO364, TYR38, LEU46, PHE62, MET111, LEU68, LEU108, ALA59, LEU359, PHE361, MET43, PRO52, ALA55, PHE98, LEU42, TYR356, LEU58 | |||
| Glycine interaction | GLY363 | |||
| 12 |
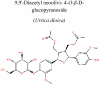
|
− 13.376 | Hydrogen bonding (side chain and backbone) | TYR356, PRO52, ARG136, THR360 |
| Polar interaction | THR63, HIE56, GLN47, THR360 | |||
| Pi-Pi stacking interaction | TYR38 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | PHE37, LEU58, MET111, LEU359, ALA59, PRO364, ALA55, PHE98, TYR356, PRO52, VAL134, LEU46, MET43, LEU67, PHE62, LEU42, LEU68, TYR38, PRO69 | |||
| Glycine interaction | GLY363 | |||
| 13 |
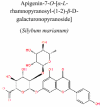
|
− 13.363 | Hydrogen bonding (side chain) | ARG136 |
| Polar interaction | GLN47, THR360, HIE56, THR63 | |||
| Salt bridge interaction | ARG136 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | LEU42, PHE62, ALA59, LEU58, PRO52, ALA55, PHE98, TYR356, MET43, LEU359, MET111, PRO364, LEU108, LEU68, TYR38, LEU46, PHE37 | |||
| Glycine interaction | GLY363 | |||
| 14 |
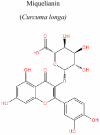
|
− 13.052 | Hydrogen bonding (side chain and backbone) | TYR356, ARG136, ALA55 |
| Polar interaction | GLN47, HIE56, THR360, THR63 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | ALA59, PHE62, LEU58, ALA55, PRO364, TYR38, LEU108, MET111, LEU68, LEU359, PHE98, TYR356, VAL134, PRO52, MET43, LEU42, LEU46 | |||
| Glycine interaction | GLY363 | |||
| 15 |
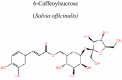
|
− 12.986 | Hydrogen bonding (side chain and backbone) | TYR38, ARG136, ALA55, PRO52 |
| Polar interaction | HIE56, GLN47, THR360 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | PRO69, TYR38, LEU68, LEU46, PHE62, ALA59, LEU58, PHE98, ALA55, TYR356, PRO52, VAL134, MET43, LEU359, PRO364, LEU42, | |||
| Glycine interaction | GLY363 | |||
| Charged (nagative) ionic interaction | GLU53 | |||
| 16 |
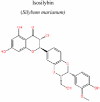
|
− 12.886 | Hydrogen bonding (side chain) | TYR356, ARG136 |
| Polar interaction | THR360, HIE56, GLN47 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | PRO69, LEU68, PRO364, PHE62, TYR38, LEU359, ALA99, ALA55, PHE98, TYR356, TYR347, VAL143, VAL134, PRO52, LEU46, LEU42, MET43 | |||
| Glycine interaction | GLY363 | |||
| Pi-Pi stacking interaction | TYR38 | |||
| 17 |
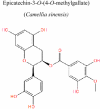
|
− 12.822 | Hydrogen bonding (side chain and backbone) | ARG136, THR360 |
| Polar interaction | HIE56, THR360, GLN47, THR63 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | LEU58, LEU46, MET43, ALA59, TYR356, ALA55, VAL134, PRO52, PHE361, LEU359, PHE98, PRO364, MET111, PHE62, LEU68, TYR38, LEU42 | |||
| Glycine interaction | GLY363 | |||
| 18 |
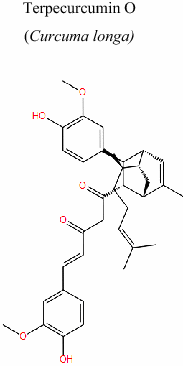
|
− 12.751 | Hydrogen bonding (side chain and backbone) | ARG136, PRO52 |
| Polar interaction | THR63, HIE56, GLN47, THR360 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Charged (negative) ionic interaction | GLU53 | |||
| Hydrophobic interaction | LEU67, MET43, LEU46, ALA59, LEU359, MET111, PHE98, ALA55, VAL134, PRO52, TYR356, PHE62, TYR38, PRO69, PRO364, LEU68, LEU42, LEU58 | |||
| Glycine interaction | GLY363 | |||
| Pi-Pi stacking interaction | TYR38 | |||
| 19 |
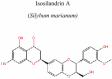
|
− 12.724 | Hydrogen bonding (side chain) | ARG136, TYR356 |
| Polar interaction | THR360, HIE56, GLN47 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Pi-Pi stacking interaction | PHE62 | |||
| Hydrophobic interaction | LEU58, TYR38, PRO364, LEU68, ALA59, LEU359, ALA55, PHE98, TYR356, VAL143, VAL134, PRO52, MET43, LEU46, PHE62, LEU42 | |||
| Glycine interaction | GLY363 | |||
| 20 |
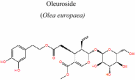
|
− 12.688 | Hydrogen bonding (side chain) | TYR38, ARG136 |
| Polar interaction | THR360, GLN47, HIE56, THR63 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Charged (negative) ionic interaction | GLU53 | |||
| Pi-Pi stacking interaction | PHE62 | |||
| Hydrophobic interaction | LEU42, TYR38, PHE62, MET43, TYR356, TYR147, VAL134, VAL143, PRO52, ALA55, PHE98, LEU359, MET111, LEU68, ALA59, PRO364, LEU58, LEU46 | |||
| Glycine interaction | Gly363 | |||
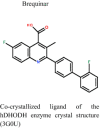
|
− 10.449 | Hydrogen bonding (side chain) | ARG136 | |
| Polar interaction | GLN47, HIE56, THR360 | |||
| Pi-Pi stacking interaction | TYR38, TYR356 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Hydrophobic interaction | PRO69, PHE62, PRO364, LEU68, LEU46, LEU359, MET43, ALA55, PRO52, VAL134, VAL143, PHE98, TYR356, LEU58, ALA59, TYR38, LEU42 | |||
| Glycine interaction | GLY363 | |||
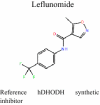
|
− 7.649 | Hydrogen bonding (side chain) | ARG136 | |
| Polar interaction | GLN47, HIE56, THR360, THR63 | |||
| Charged (positive) ionic interaction | ARG136 | |||
| Pi-Pi stacking interaction | ARG136 | |||
| Glycine interaction | GLY363 | |||
| Hydrophobic interaction | ALA59, LEU359, VAL134, PRO52, TYR356, ALA55, PHE98, MET43, LEU46, PHE62, PRO364, TYR38, LEU68, MET111 |
