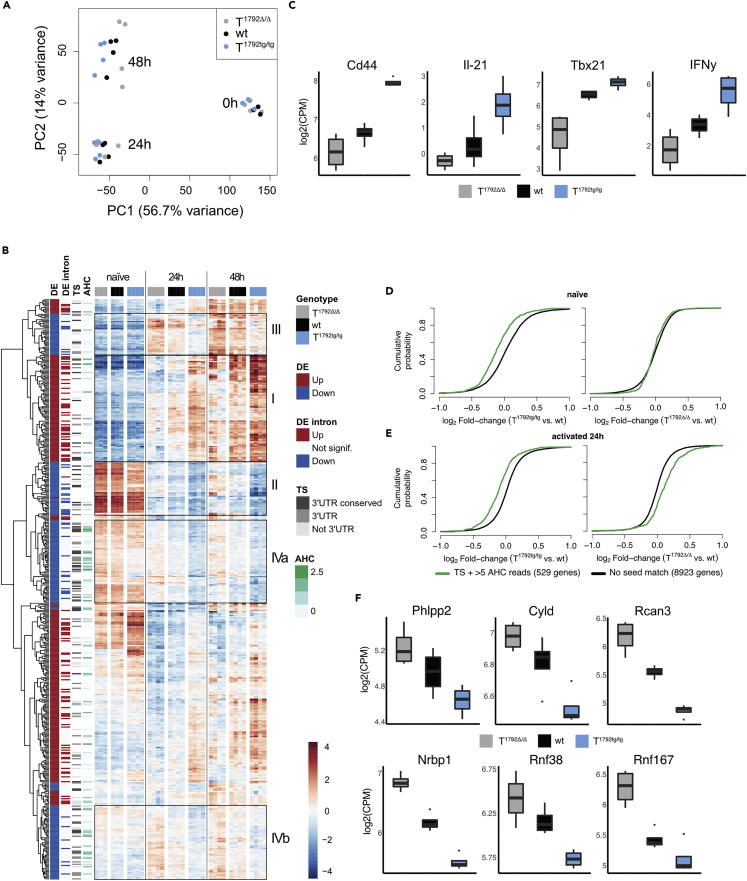
Figure 4.
miR-17∼92 shapes the transcriptome after CD4+ T cell activation
Naive CD4+ T cells from T1792Δ/Δ (gray), wt (black) and T1792tg/tg (light blue) were activated with plate-bound anti-CD28 and anti-CD3 for 0, 24, and 48 h. Total RNA was extracted for bulk RNA sequencing.
(A) PCA (PC1 vs. PC2) based on the 25% most variable genes.
(B) Hierarchical clustering of the set of genes selected with abs(log2FC) > 1 & adj.P.Val<0.001 in the T1792tg/tg vs. T1792Δ/Δ comparison at 24 h. The heatmap displays the centered log of counts per million, with blue indicating low and red indicating high expression. Annotations: “DE” indicates the fold change direction, “DE intron” indicates if significant changes are observed in EISA analysis, “TS” indicates presence (gray) or absence (blank) of a seed match and its location and “AHC” indicates the 3′UTR signal intensity in HITS-CLIP data. Boxes I–IVb designate gene clusters.
(C) Examples of genes in cluster I.
(D and E) Genome-wide transcriptome analysis presented as the log2 value of the gene-expression ratio for each gene versus the cumulative fraction of all log2 ratios in naive (D) and 24 h activated (E). Shown is the miR-17 seed family for T1792tg/tg vs. wt and T1792Δ/Δ vs. wt comparisons. Black curve: all genes without a seed match and ≤5 AHC reads; green: subset of genes with a seed sequence for the seed family and >5 reads in the AHC.
(F) Examples of genes defined as empirically validated miR-17∼92 targets. log2 RNA expression level in activated CD4+ T cells, numbers correspond to FDR<0.05. Boxplot representing values distribution over minimum and maximum values, median, 25th and 75th percentiles.