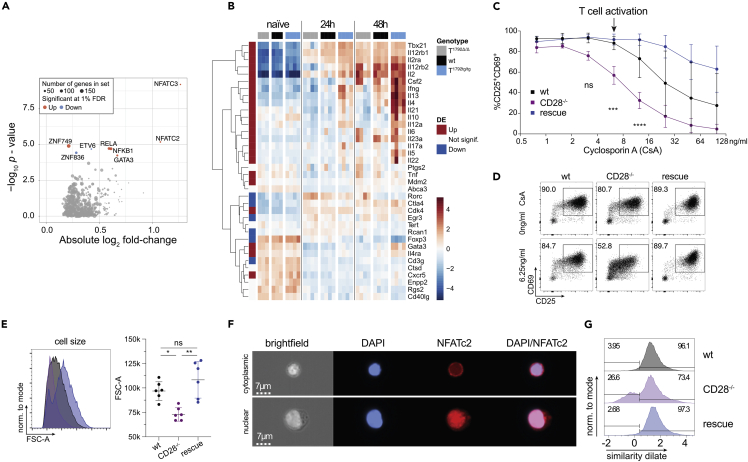
Figure 5.
miR-17∼92 promotes the NFAT and NF-κB pathways in CD4+ T cells
(A and B) Dataset from Figure 4. Naive CD4+ T cells from T1792Δ/Δ (gray), wt (black), and T1792tg/tg (light blue) were activated with plate-bound anti-CD28 and anti-CD3 for 0, 24, and 48 h. Total RNA was extracted for bulk RNA sequencing. (A) Volcano plot showing the absolute log2 fold change and -log10 p value from regulon analysis. A threshold of 1% FDR was applied. Dot size indicates the number of genes within each regulon and colors indicate fold change direction. (B) Heatmap of genes under NFATC2_D and NFATC3_D regulons plus several known activated genes in CD4+ T cells (Il10, Il12a, Il6, Rorc, Il23a). Hierarchical clustering was applied on genes. Centered log of counts per million is displayed, with blue indicating low and red indicating high expression.
(C) CD4+ wt (black), CD28−/− (purple), and rescue (dark blue) cells were activated for 48 h in the presence of increasing concentrations of cyclosporin A (CsA) as indicated and stained for CD25 and CD69. Shown are 2 independent experiments, error bars represent means ± SD. Tukey’s multiple comparison, p values: ∗∗<0.002, ∗∗∗∗<0.0001 refer to the difference between CD28−/− and wt.
(D) representative FACS plots of CD25/CD69 expression in viable CD4+ T cells activated for 48 h with no or 6.25 ng/mL CsA.
(E) influence of 6.25 ng/mL CsA on blasting (FSC-A of the lymphocyte gate) of viable CD4+ cells.
(F) Imagestream analysis of CD4+ T cells, activated for 48 h in presence of 6.25 ng/mL CsA; DAPI (blue) and NFATc2 (red) staining. Examples for cytoplasmic (top) and nuclear (bottom) NFATc2 in a CD28−/− sample.
(G) histograms of the similarity dilate indicative of the co-localization of NFATc2 and DAPI signals; gates indicate the nuclear (high similarity dilate) and the cytoplasmic population (low similarity dilate).
See also Figures S5 and S6.