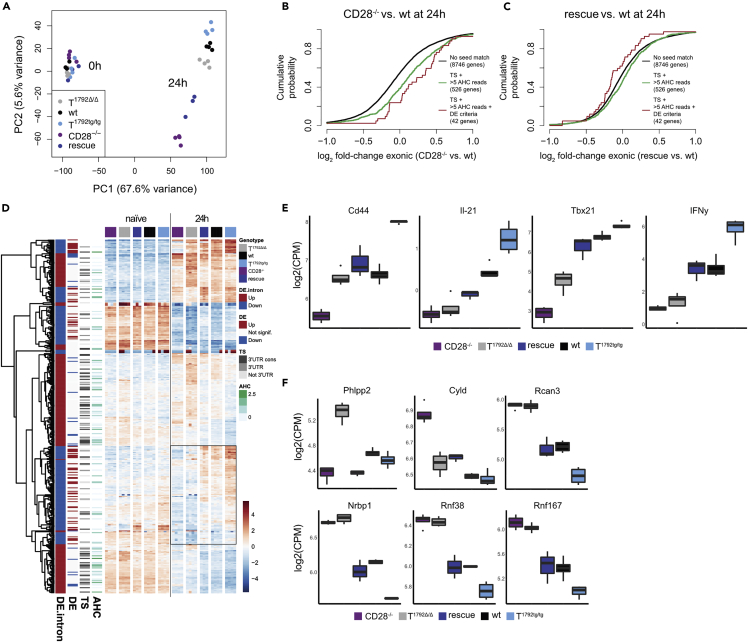
Figure 6.
miR-17∼92 is necessary for repression of a subset of genes during T cell activation
CD4+ T cells from T1792Δ/Δ (gray), wt (black), T1792tg/tg (light blue), CD28−/− (purple) and rescue (dark blue) mice were activated for 24 h. Total RNA was extracted for sequencing.
(A) PCA (PC1 vs. PC2) based on the 25% most variable genes.
(B and C) (B) Genome-wide transcriptome analysis, presented as the log2 value of the gene-expression ratio for each gene versus the cumulative fraction of all log2 ratios. Shown are the contrasts between activated samples separated by the miR-17 seed family for the comparison CD28−/− vs. wt (B) and rescue vs. wt (C). Black curve: genes without a seed match, ≤5 AHC reads, and no differential expression in the second RNA sequencing. Green: genes with a conserved binding site for the miR-17 seed family (TS) and >5 reads in the AHC, red: genes with a conserved binding site for the miR-17 seed family (TS), >5 reads in the AHC and differential expression in the second RNA sequencing dataset.
(D) Hierarchical clustering of the set of genes selected with abs(log2FC) > 1 & adj.P.Val<0.001 in the T1792tg/tg vs. T1792Δ/Δ comparison at 24 h. The heatmap displays the centered log of counts per million, with blue indicating low and red indicating high expression.
(E) Examples of transcripts that were restored in “rescue” compared to CD28−/− T cells, i.e. contained in the box (D).
(F) Examples of direct miR-17∼92 targets. E, F: log2 mRNA expression (counts per million) in activated CD4+ T cells; numbers correspond to FDR<0.05. Boxplot representing values distribution over minimum and maximum values, median, 25th and 75th percentiles.
See also Figure S7.