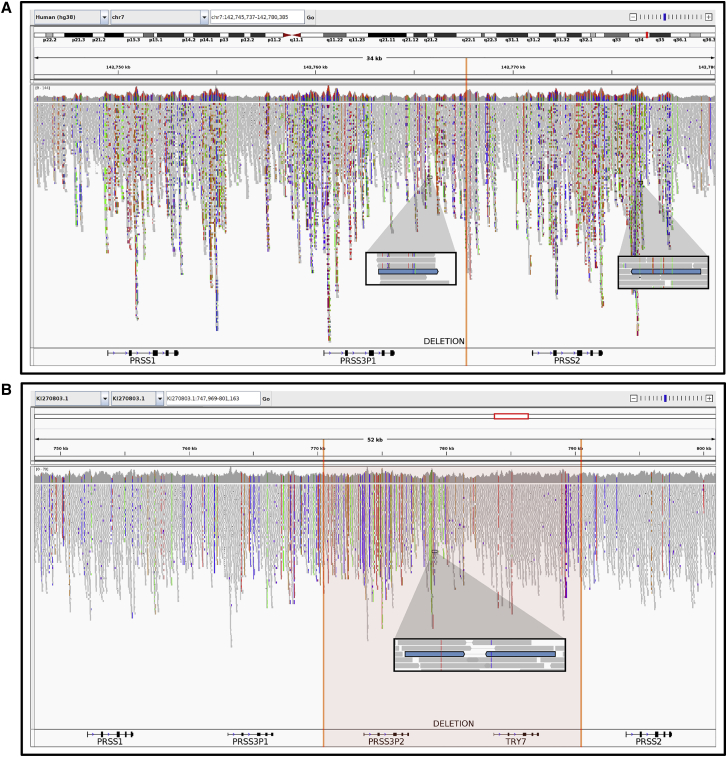
Figure 6.
Demonstration of reference bias induced by 20-kb deletion polymorphism and correction using alternative contig
GRCh38 chr7 reference sequence excludes 20-kb present in the sequenced individual (orange vertical lines); reads that would align in this region generate spurious mappings to nearby homologous positions.
(A)IGV visualization of 10XG reads aligned to the GRCh38 chr7 reference chromosome. Regions of higher coverage and an excessive variation are produced. A highlighted read pair maps kilobases apart, downstream of PRSS3P1 and PRSS2.
(B)IGV visualization of 10XG reads aligned to the KI270803.1 alternative contig. Alignments produce more uniform coverage and less variation with respect to the reference. The same highlighted read pair maps with a reasonable insert size, downstream of PRSS3P2. The use of KI270803.1 instead of the GRCh38 chr7 reference sequence produces more accurate alignments for individuals carrying the 20-kb absent from the reference sequence.