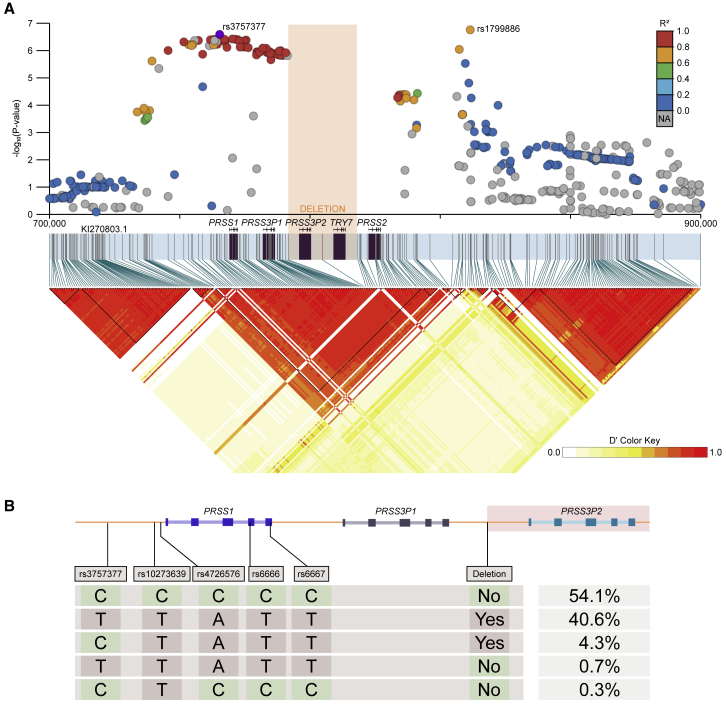
Figure 8.
Linkage and haplotype structure at the chr7q35 trypsinogen locus
(A)LD matrix calculated from 10XG phased calls; deletion allele is denoted by orange rectangle. Haplotype blocks are drawn as black triangles, and all five trypsinogen paralogs are located within a single block (KI270803.1:737033–802909). Meconium ileus GWAS summary statistics lifted from GRCh37 to KI270803.1 are shown, R2 with respect to rs3757377.
(B)Four SNPs in the same LD block as rs3757377, phased with the deletion polymorphism. SNPs include a common pancreatitis risk allele (rs10273639),18 a PRSS1 promoter SNP (rs4726576) that alters expression of a reporter gene in mice,19 and two synonymous PRSS1 variants (rs6666 and rs6667). Five unique haplotypes are observed in 10XG data, and the frequencies are shown as a percentage. The two major haplotypes account for 94.7% of the observed data. The two rarest haplotypes (1% of the observed data) were supported by manual inspection in IGV.