Figure 1.
Proteomic profiling of sacsin KO cells
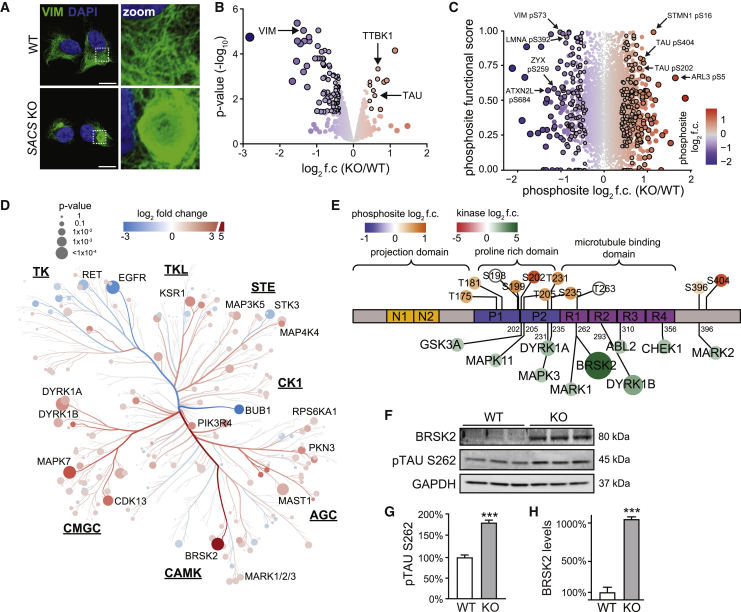
(A) Representative confocal images of control (WT) and sacsin KO SH-SY5Y neuroblastoma cells immunostained for the intermediate filament protein vimentin. Scale bars, 10 μm.
(B) Global proteomic profiling of sacsin KO SH-SY5Y cells. Significance cutoffs: p < 0.05 and log2 fold change (f.c.) ±0.4, denoted by black outline.
(C) Functional analysis of altered phosphosites in sacsin KO cells. y axis is the functional score assigned by Ochoa et al. (2020), with higher scores reflecting increased effects on fitness. Dot color and size reflect log2 f.c. Black outlines label phosphosites with p < 0.05 and log2 f.c. ±0.4.
(D) Phylogenetic tree of the kinome in sacsin KO cells. Color indicates log2 f.c. of kinase abundance, size indicates −log10 p value. Underlined abbreviations refer to phylogenetically related kinase families.
(E) Protein map of tau isoform 2 (2N4R). Phosphosites identified in phosphoproteomic profiling are labeled above diagram. Tau kinases identified in the kinome profiling are listed below, indicating validated phosphosites. Colored circles correlate with log2 f.c. of differentially expressed phosphosites or kinases.
(F–H) Western blot and quantification for BRSK2, and the BRSK2 target residue pTAU S262. n = 3, SEM, Student’s t test, ∗∗∗p < 0.001.