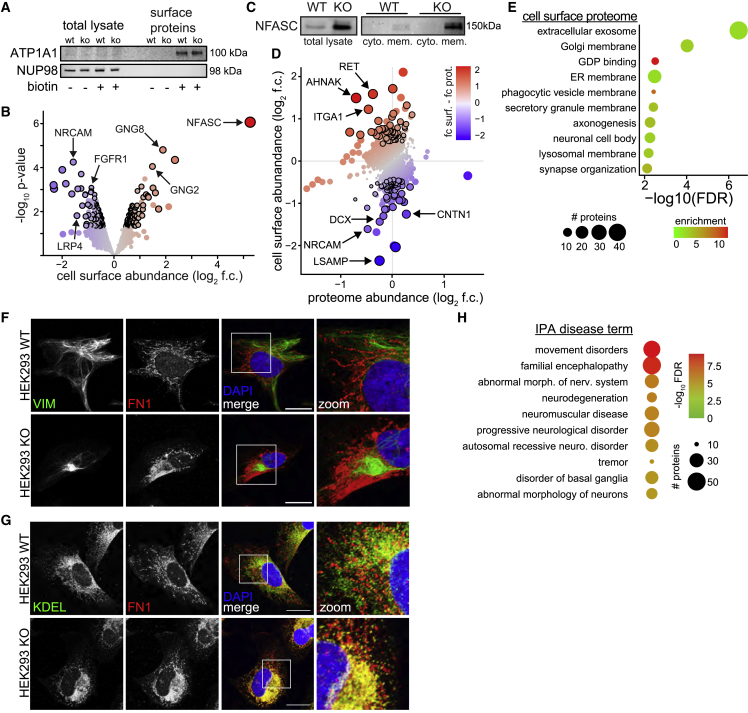
Figure 5.
The loss of sacsin affects the localization of cell adhesion proteins
(A) Western blot of cell surface protein purification, illustrated by the membrane protein ATP1A1, and the nuclear pore protein NUP98. After purification ATP1A1 is detectable only in conditions that were treated with biotin, and NUP98 is no longer detected, suggesting labeling specificity and enrichment of cell surface proteins.
(B) Mass spectrometry of cell surface proteins in sacsin KO SH-SY5Y cells. Significance cutoffs: p < 0.05 and log2 fold change (f.c.) ±0.4, denoted by black outline. Proteins which pass these cutoffs, but were also detected in biotin negative controls, were not considered for downstream analysis (Table S1).
(C) Western blot of NFASC in total lysate (left), and fractionated cytoplasmic or membrane fractions in WT and sacsin KO cells.
(D) Levels of proteins detected in both cell surface and proteomic datasets. Proteins are colored by the disparity between these two datasets (f.c. surface and f.c. proteome), with red indicating more, and blue less membrane abundance relative to total protein levels. Black outlines are proteins with p < 0.05, log2 f.c. ± 0.4 in the surface dataset.
(E) GO term analysis of proteins differentially localized in membrane of sacsin KO cells (p < 0.05, log2 f.c. ±0.4).
(F and G) Confocal images for fibronectin and vimentin (F) and ER marker KDEL (G) in WT and sacsin KO HEK293 cells. Scale bars, 10 μm.
(H) Disease enrichment analysis with Ingenuity Pathway Analysis of differentially localized surface proteins (p < 0.05, log2 f.c. ±0.4).