FIGURE 7.
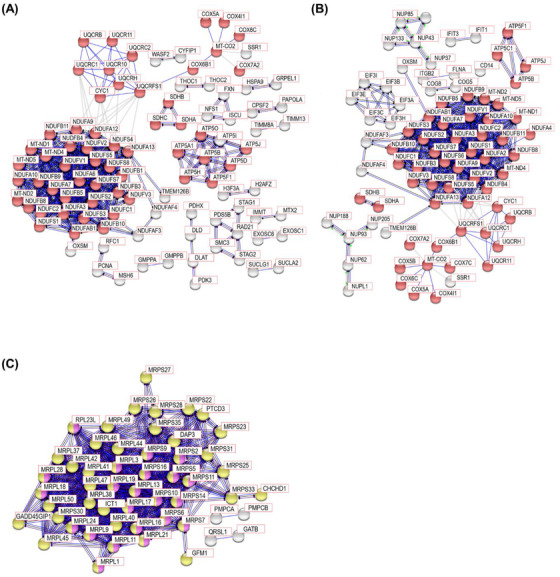
STRING PPI analysis for network connectivity of the modules from WGCNA enriched with Parkinson's disease, mitochondria translation, and ribosome pathways. STRING PPI analysis with the proteins in (A) the M12 module generated by WGCNA of PSP and HC, (B) the M6 module generated by WGCNA of PSP and PD, and (C) the M11 module generated by WGCNA of PSP and HC. In the M12 module from PSP and HC, the network contains 422 nodes with 501 edges. Only experimental data was used for the active interaction source with 0.9 highest confidence threshold of a minimum required interaction score (average node degree: 2.37, average local clustering coefficient: 0.193, and PPI enrichment p value <1.0 ×10−16). In the M6 module from PSP and PD, the network contains 997 nodes with 702 edges. Only experimental data was used for the active interaction source with 0.9 highest confidence threshold of a minimum required interaction score (average node degree: 1.41, average local clustering coefficient: 0.203, and PPI enrichment p value <1.0 ×10−16). In the M11 module from PSP and HC, the network contains 187 nodes with 542 edges. Only experimental data was used for the active interaction source with 0.9 highest confidence threshold of a minimum required interaction score (average node degree: 5.8, average local clustering coefficient: 0.219, and PPI enrichment p value <1.0 ×10−16). The red, yellow, and magenta nodes denote Parkinson's disease, mitochondria translation, and ribosome pathways, respectively. The grey nodes do not belong to any enriched pathways.
