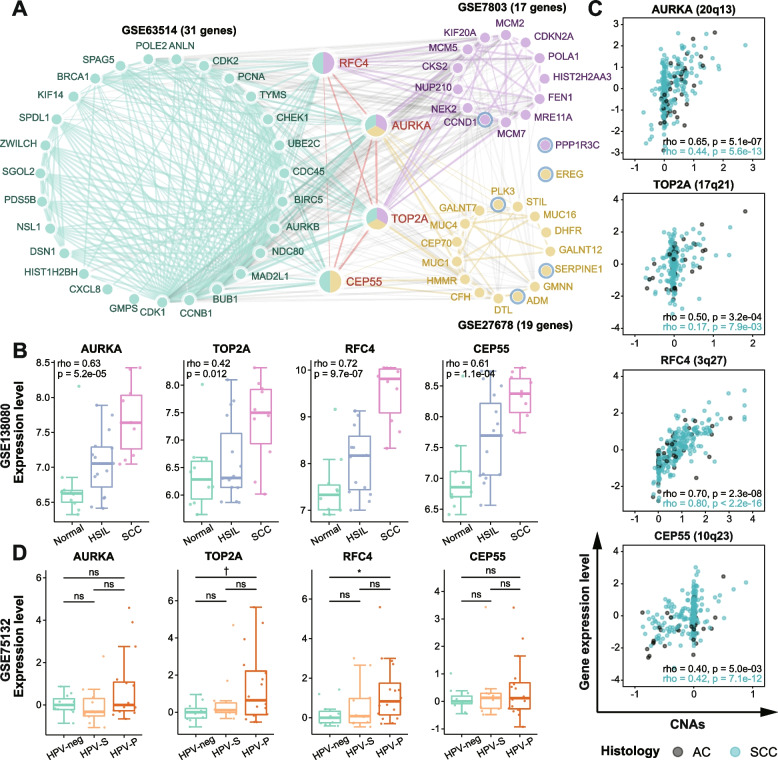
Fig. 4.
Hub genes identification and validation. A PPI network of important DEGs selected by Cytohubba. The nodes with white and blue rings denote progressively up- and downregulated genes with the development of cervical lesions (Spearman, p < 0.05). Edge thickness is proportional to the interaction score. B Boxplots showing the correlations between hub gene expression and severity of cervical lesion in GSE138080, with Spearman’s rho and p-values presented in the upper left corner. C Scatter plots showing the correlations between hub gene expression (Z score-transformed log2 (FPKM-UQ+1) values) and CNAs in TCGA-CESC dataset, with Spearman’s rho and p-values presented in the lower right corner. Adenocarcinoma (AC) samples are shown in black and squamous cell carcinoma (SCC) samples are shown in blue. D Boxplots showing hub gene expression in HPV-neg, HPV-S (HPV16 persistent infection without progression), and HPV-P (HPV16 persistent infection with progression) women from GSE75132. Statistical comparisons were performed using Wilcoxon rank-sum test. * 0.01 < p < 0.05; † p < 0.1; ns, p ≥ 0.1