Summary
Human chromosomes are pervasively transcribed, but systematic understanding of coding and long noncoding RNA (lncRNA) genome function in cell differentiation is lacking. Using CRISPR interference (CRISPRi) in human induced pluripotent stem cells, we performed dual genome-wide screens—assessing 18,905 protein-coding and 10,678 lncRNA loci—and identified 419 coding and 201 lncRNA genes that regulate neural induction. Integrative analyses revealed distinct properties of coding and lncRNA genome function, including a 10-fold enrichment of lncRNA genes for roles in differentiation compared with proliferation. Further, we applied CRISPRi perturbation coupled with single-cell RNA-seq (Perturb-seq) to obtain granular insights into neural induction phenotypes. While most coding hits stalled or aborted differentiation, lncRNA hits were enriched for the genesis of diverse cellular states, including those outside the neural lineage. In addition to providing a rich resource for understanding coding and lncRNA gene function in development, these results indicate that the lncRNA genome regulates lineage commitment in a manner fundamentally distinct from coding genes.
Keywords: CRISPR, CRISPRi, human pluripotent stem cells, iPSCs, neurodevelopment, neural induction, long noncoding RNA, genome-wide pooled screens, Perturb-seq, single-cell RNA-seq
Graphical abstract
Highlights
-
•
Dual genome-wide screens provide coding and lncRNA functional atlas in human iPSCs
-
•
Compared with coding genes, lncRNA genes are enriched for roles in neural induction
-
•
Perturb-seq reveals fundamental insights into regulation of neural induction
-
•
Interactive resource allows data exploration: danlimlab.shinyapps.io/dualgenomewide
There is limited systematic understanding of coding and lncRNA genome function in processes such as differentiation. Wu et al. performed dual genome-wide coding and lncRNA CRISPRi screens and Perturb-seq in neural induction from pluripotent stem cells, finding fundamentally distinct roles of coding and noncoding genes in this complex biological process.
Introduction
The human genome expresses thousands of genes—both coding and noncoding1,2—and many are critical to the complex processes of cell differentiation during development.3,4,5 Early in mammalian development, neural stem cells (NSCs) are produced from pluripotent stem cells by the process of neural induction.6 Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides that do not encode protein, and many are expressed in neural tissues.2,7,8 The recent evolutionary expansion of these loci has led to the hypothesis that lncRNA genes play critical roles in the development of complex organisms.8,9,10 However, unlike coding genes, far fewer lncRNA genes have been demonstrated to regulate cell biology.11 More broadly, systematic understanding of how the coding and lncRNA genomes regulate developmental processes is lacking.
Genetic screens are powerful methods for identifying genes underlying phenotypes of interest.12 The vast majority of CRISPR-based screens have focused on the protein-coding genome, typically excluding lncRNA loci. Nevertheless, these studies provide insight into principles of coding genome function by integrating screen data into a rich foundation of literature, including knowledge of physical and functional interaction networks. Although genetic screens of lncRNAs are now emerging,13,14,15 functional knowledge for this class of molecules is still primarily drawn from the study of individual lncRNAs. Genome-wide screens that integrate information from both the coding and lncRNA genomes are rare14 and have not been performed in complex contexts such as cell differentiation. Such dual genome-wide approaches can provide unique data resources to discover principles of developmental regulation.
In this work, we used functional genomics to systematically assess 18,905 coding genes and 10,678 lncRNAs for roles in human neural induction. Using dual genome-wide CRISPR interference (CRISPRi) marker-based screens, we identified 419 protein-coding and 201 lncRNA genes that regulated the production of NSCs from induced pluripotent stem cells (iPSCs). The scale and design of this resource enabled integrated analyses and the discovery of general properties of coding and lncRNA genome function. To obtain deeper insights into the biology of these regulators, we applied this resource to perform a CRISPRi perturbation coupled with single-cell RNA sequencing (RNA-seq), known as Perturb-seq.16,17,18,19,20 Collectively, these systematic studies revealed fundamental insights about the unique developmental roles of the coding and lncRNA genomes at a level that is challenging to ascertain by the study of individual genes.
Results
Dual genome-wide CRISPRi screens identify coding and noncoding genes regulating neural induction
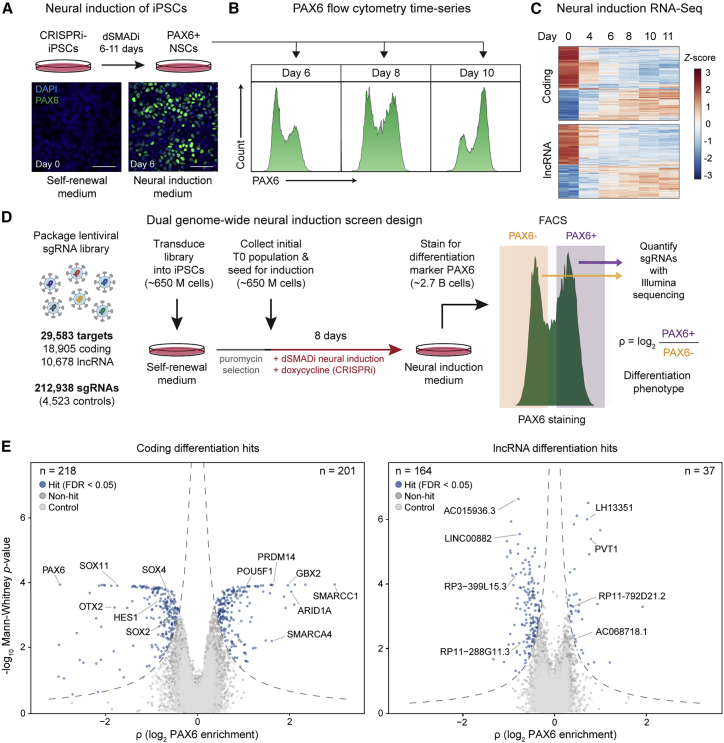
An early step toward brain development is neural induction from pluripotent stem cells. Using dual SMAD inhibition (dSMADi),6,21 we induced NSCs from iPSCs that express dCas9-KRAB (CRISPRi-iPSCs) under doxycycline-inducible control (Figure 1A). The induction of NSCs was progressive over time, which we characterized by flow cytometry analysis of the canonical marker PAX6 (Figure 1B) and RNA-seq of polyadenylated and total RNA at multiple time-points (0–11 days). Many thousands of coding and noncoding genes were dynamically expressed over the course of neural induction (Figure 1C; Table S1).
Figure 1.
Dual genome-wide CRISPRi screens identify coding and noncoding genes regulating neural induction
(A) Neural induction of CRISPRi-iPSCs by dual SMAD inhibition, stained for PAX6 protein, a canonical marker of NSCs. Scale bar, 50 μm.
(B) Progressive increase of PAX6+ NSCs over time during neural induction, analyzed by flow cytometry. Distinct peaks of PAX6+ and PAX6− were present at day 8, which was selected as the screen endpoint.
(C) Heatmap showing Z-scaled gene expression of differentially expressed coding and lncRNA transcripts during neural induction.
(D) Overview of pooled, marker-based genome-wide CRISPRi screen design for assessing 18,905 coding genes and 10,678 lncRNA targets in the regulation of neural induction. Each class was targeted with ∼100,000 sgRNAs, with five sgRNAs/transcriptional start sites (TSSs) for coding genes and 10 sgRNAs/TSSs for lncRNA genes. Top and bottom thirds of the PAX6 fractions were sorted during FACS for next-generation sequencing.
(E) Volcano plots of screen results for coding and lncRNA genes, with x axis showing screen phenotype rho (ρ) value (log2 enrichment in PAX6+/PAX6− fractions) and y axis showing −log10 p value. Blue dots show hits (FDR < 0.05), dark gray dots show non-hits, and light gray dots show non-targeting controls.
We applied the transcriptomic data to inform the assembly of a dual genome-wide library (STAR Methods) containing published, validated CRISPRi single-guide RNAs (sgRNAs) targeting human coding (hCRISPRi-v2)22 and lncRNA (CRiNCL) genes.13 These sgRNAs were selected based on RNA-seq expression during neural induction and were designed in prior studies using an algorithm that incorporates nucleosome-positioning and FANTOM cap analysis of gene expression data, with off-target activity filtering.13,22 We included a total of 212,938 sgRNAs (with 4,523 non-targeting controls) against 29,583 targets, covering 18,905 coding (five sgRNAs/target) and 10,678 lncRNA genes (10 sgRNAs/target).
We conducted the dual genome-wide screens using CRISPRi-iPSCs with PAX6 staining as the readout for neural induction (Figure 1D). We selected day 8 of neural induction as the endpoint, when both PAX6+ and PAX6− populations were present (Figure 1B), enabling the identification of hits that either increase or decrease this marker of neural induction. After sequencing the assembled library to ensure uniform distribution, we packaged lentivirus and transduced ∼650 million CRISPRi-iPSCs in two biological replicates. Cells were propagated in self-renewal media under puromycin selection until reaching >80% sgRNA positivity, detected by co-expressed blue fluorescent protein (BFP).
We maintained the screens at >1,000× sgRNA coverage per replicate over the course of neural induction. Time zero (T0) aliquots were collected to assess initial sgRNA abundance. After 8 days of neural induction and dCas9-KRAB expression, cells were harvested and quantified. Approximately 2.7 billion cells in total were fixed, permeabilized, and stained with antibodies against PAX6 for fluorescence-activated cell sorting (FACS) into PAX6+ and PAX6− fractions (top and bottom thirds; Figure 1D, right). The abundance of sgRNAs in each fraction were quantified by PCR amplification followed by Illumina sequencing.
The differentiation phenotype rho (ρ; log2 enrichment ratio of normalized sgRNA abundance in PAX6+ versus PAX6− fraction) was calculated for all targets and non-targeting controls (Figure 1D, right). This ρ value, used in other marker-based studies, represents the log2 fold change of each sgRNA in the positive fraction relative to the negative fraction.13,16,23 Negative values of ρ indicate that the sgRNA decreased neural induction (e.g., knockdown of pro-neural factors), while positive ρ values indicate that the sgRNA promoted the development of PAX6+ cells (e.g., knockdown of pluripotency factors). Independent replicates were correlated (Figure S1A) and non-targeting control sgRNAs produced ρ values centered around zero, as expected (Figure S1B). More than 99% of sgRNAs met a threshold of >100× coverage (with 97% with >500×) providing sufficient data for all 29,583 targets, with 94% of targets having all designed sgRNAs represented. After applying an empirical false discovery rate (FDR) of 0.05, exclusion of sgRNAs targeting multiple loci and gene “neighbor hits” (STAR Methods), we identified 419 protein-coding and 201 lncRNA genes that altered the production of PAX6+ NSCs (Figure 1E).
Since each hit was targeted by multiple sgRNAs, we assessed whether these sgRNAs were in agreement by calculating the fraction of sgRNAs in the same direction as the hit. Hits showed a very high median concordance of 1 (indicating that all sgRNAs had the same effect) while those targeting non-hits had a median concordance of 0.5 (indicating random chance) (Figure S1C). Additionally, given the large scale of the screen, we estimated hit identification performance at smaller scales by downsampling the raw data for precision-recall analysis. At 10% downsampling (∼100× coverage), performance was poor (<40% of hits identified). This improved substantially at 200× and 500× coverage, where >70% and >80% of hits were identified, respectively (Figure S1D). Thus, the comprehensive scale of the dual genome-wide screens provides an unparalleled glimpse into this early differentiation process.
Validation of genome-wide screen results
Of the 18,905 coding genes screened, PAX6 itself was expectedly the highest scoring negative hit (Figure 1E) with ρ = −3.01, representing an 88% reduction in PAX6+ cells by FACS. We also observed numerous examples of hits with expected positive or negative impact on neural induction. For instance, pro-pluripotency factors (POU5F1/OCT4, GBX2, SMARCC1, PRDM14) were positive hits while genes with known neurodevelopmental roles (SOX2, SOX4, SOX11, HES1, OTX2) were negative hits. Protein-protein network analysis revealed enrichment for known functional interactions among coding gene hits, such as those of the BRG1/BRM-associated factor (BAF) chromatin remodeling complex, the Polycomb repressive complex (PRC), and signaling pathways critical to neurodevelopment such as NOTCH (Figures S2A and S2B). Furthermore, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of coding hits revealed enrichment for processes important in early development (Figure S2C). Thus, our screen recovers a large number of genes known to function in complexes and pathways important for neural induction.
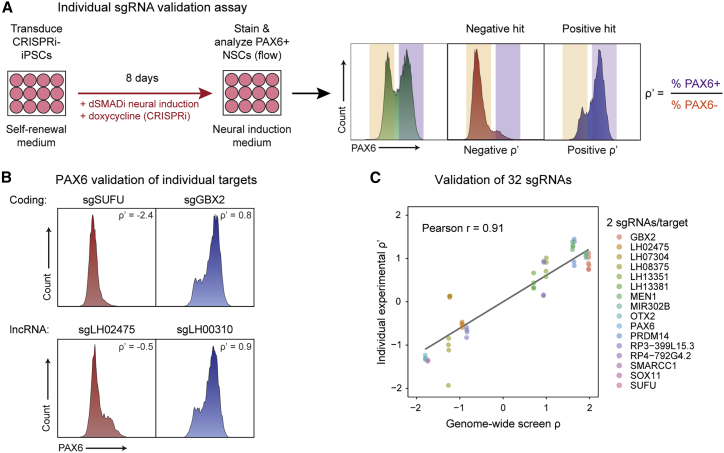
To experimentally validate screen results, we selected 16 hits and targeted each with two independent sgRNAs (32 different sgRNAs in total covering each of the hit subcategories, i.e., coding/lncRNA, positive/negative, with two biological replicates per sgRNA). These sgRNAs were individually transduced into iPSCs, and, after 8 days of neural induction and CRISPRi, the cells were analyzed by PAX6 staining via flow cytometry (Figure 2A). Individual sgRNAs targeting both coding and lncRNA genes showed phenotypes matching their screen phenotypes (Figure 2B). Collectively, quantitative results from individual experiments were highly correlated with the screen ρ phenotype (Pearson r = 0.91, Figure 2C), providing experimental validation to the hits identified in the screen.
Figure 2.
Experimental validation of genome-wide screen results
(A) Diagram of arrayed, individual sgRNA validation assay using CRISPRi-iPSCs with exemplar PAX6 flow cytometry histograms for negative and positive screen hits.
(B) Histograms showing PAX6 flow cytometry staining of validated positive and negative hits for both coding and lncRNA classes.
(C) Validation scatterplot of 32 sgRNAs targeting positive and negative hits of both coding and lncRNA classes (16 total targets × 2 replicates × 2 sgRNAs), with x axis showing the genome-wide screen ρ and y axis showing the individual validation ρ′, which were strongly correlated (Pearson r = 0.91).
See also Figure S2.
lncRNA genes are enriched for roles in promoting neural induction
Similar numbers of coding gene hits exerted positive (52%) and negative (48%) effects on neural induction, and this slight bias was not significant (permutation test p = 0.27; Figure 3A, left). In contrast, the majority (87%) of lncRNA hits identified were negative hits, and this enrichment in the lncRNA hit distribution was highly significant (permutation test p < 1 × 10−6; Figure 3A, right). These results indicate that lncRNA hits were enriched for functions that normally promote neural induction.
Figure 3.
lncRNA genes are enriched for roles in promoting neural induction
(A) Distributions of genome-wide ρ values for coding and lncRNA hits in PAX6 marker-based differentiation screens. ∗p < 1 × 10−6 by permutation test.
(B) Distributions of genome-wide γ values for coding and lncRNA hits in growth-based proliferation screens. ∗p < 1 × 10−6 by permutation test.
(C) Scatterplots showing both differentiation (ρ) and proliferation (γ) phenotypes for all screened genes, with hits colored by their primary phenotype (differentiation, blue; proliferation, orange; or dual, purple) and non-hits shown in gray.
(D) Venn diagrams showing relative breakdown of coding and lncRNA hits by their primary phenotypes, drawn to scale.
(E) Ratio of differentiation to proliferation hits for coding genes and lncRNAs in neural induction. ∗p < 1 × 10−6 by permutation test.
During development, cell division can have important effects on differentiation. To investigate the effects of proliferation during neural induction, we compared the total sgRNA abundance in the PAX6+ and PAX6− fractions at day 8 (final abundance) with the initial sgRNA abundance in samples collected at the beginning of the screen (Figure S3A). This enabled calculation of the growth enrichment index gamma (γ; negative values indicate a decrease in proliferation; positive values indicate an increase), which we directly validated in a separate screen without FACS (STAR Methods; Figure S3B).
We identified 730 coding gene hits and 24 lncRNA gene hits that altered cell proliferation during neural induction (Table S2). As expected, coding gene hits included numerous cell cycle, apoptosis, and other essential genes (e.g., CDC20, CDT1, TP53, MDM2, TOP2A, BAX) (Figure S3C). Proliferation hits were strongly enriched for GO terms relating to essential biological processes, including ribosome biogenesis and DNA helicase activity (Figure S3D). As a group, coding hits were biased toward negative proliferation hits (Figure 3B), consistent with previous studies of essential genes.24,25,26
Integrated analyses of both differentiation and proliferation effects (Figure 3C) revealed that the majority (91%) of hits produced a single phenotype (i.e., differentiation or proliferation, but not both). Of the 1,258 hits across both coding and lncRNA genomes, only 9% of hits had dual phenotypes (Figure 3D). For example, knockdown of the dual hit POU5F1/OCT4 increased differentiation (positive p; Figure 1E) and decreased proliferation (negative γ; Figure S3C), consistent with its role in maintaining both pluripotency and self-renewing stem cell divisions.27,28
Notably, the coding and lncRNA genomes differed vastly in their propensity for differentiation and proliferation phenotypes. Among coding genes, there were only half as many differentiation hits compared with proliferation hits (permutation test, p < 1 × 10−6). In stark contrast, among lncRNA genes, differentiation hits outnumbered proliferation hits by over 9-fold (permutation test, p < 1 × 10−6; Figure 3E). These differences in differentiation versus proliferation ratios highlight the unique roles of these two aspects of the genome in regulating cell biology. Overall, these integrated analyses of the dual genome-wide screen results indicate that the lncRNA genome is far more specialized for roles in promoting neural induction compared with the coding genome.
Distinct transcriptomics and epigenomics of coding and lncRNA gene hits
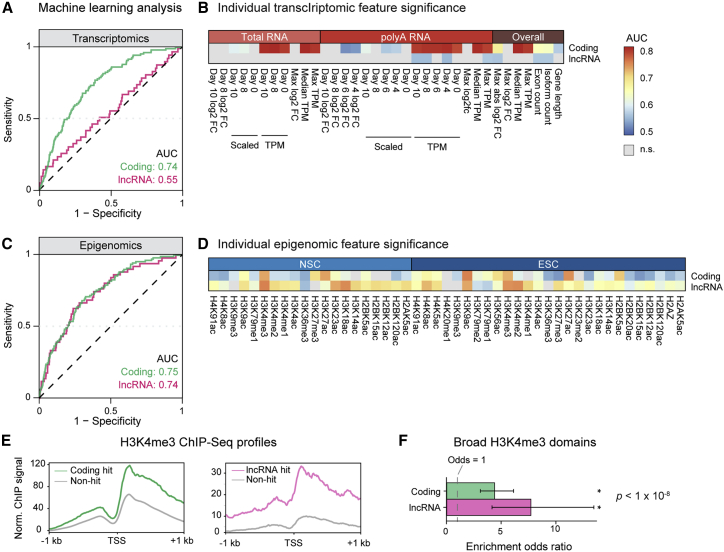
We next leveraged the screen data to identify transcriptomic and epigenomic properties that distinguish hits from non-hits. A priori, we hypothesized that differential expression would be predictive of hits. For example, negative hits may have expression patterns similar to PAX6 (high in NSCs, low in stem cells), whereas positive hits may have the expression pattern of POU5F1/OCT4 (high in stem cells, low in NSCs). However, examination of individual genes revealed both negative (e.g., PAF1) and positive (e.g., SMARCE1) hits with stable expression throughout neural induction (Figure S4A). To systematically assess the relationships of transcriptomic and epigenomic data to screen phenotypes, we turned to a machine learning approach.
To provide transcriptomic features for this analysis, we used our neural induction RNA-seq time-series data. For each target in the screen, we determined the gene expression (transcripts per million [TPM]), fold change at each time-point relative to day 0, maximum expression, maximum fold change, and scaled expression (Z score representing relative change over time). For epigenomic features, we used data from the Roadmap Epigenomics project that profiled 27 histone marks in human embryonic stem cells (ESCs) undergoing dual SMAD inhibition neural induction similar to that performed in our screen.29 Specifically, an individual epigenomic feature would be the level of a histone mark in the stem cell and NSC stages. For all coding and lncRNA gene promoters, we quantified the levels of the histone marks at these stages.
To compare the overall ability of transcriptomic and epigenomic data to discriminate hits from non-hits, we constructed machine learning classifiers and analyzed the area under the curve (AUC) of the receiver operating characteristic (ROC) (Figure S4B). While transcriptomic data were able to classify coding hits (mean AUC, 0.74), these data performed poorly for lncRNA hits (mean AUC, 0.55) overall and in a bootstrapped analysis of individual features (Figures 4A and 4B). The median expression level of coding hits (50.4 TPM) was more than 4-fold higher than that of non-hits (11.7 TPM), whereas the expression level difference of lncRNA hits (0.7 TPM) and non-hits (0.4 TPM) was smaller and less significant (Figure S4C, left). Differential expression differences (maximum absolute fold change) were also more associated with coding hits than lncRNA hits (Figure S4C, right). Furthermore, in an analysis of temporal expression dynamics, coding hits were associated with certain expression patterns, but lncRNA hits were not enriched for any pattern (Figures S4D and S4E). Thus, common transcriptional heuristics used to predict the biological activity of coding genes—such as expression level or temporal pattern—do not apply to lncRNA genes.
Figure 4.
Machine learning analyses reveal distinct transcriptomic and epigenomic properties of coding and lncRNA hits
(A) Representative ROC curves for transcriptomic data in classifying coding and lncRNA hits versus non-hits. Selected curves were within 1% of the mean AUC of 1,000 training/validation trials.
(B) Heatmaps showing AUC values for individual transcriptomic features for classifying coding and lncRNA hits versus non-hits. Statistical significance determined at the 99% confidence level from 1,000 bootstraps; non-significant features denoted in gray.
(C) Representative ROC curves for epigenomic data in classifying coding and lncRNA hits versus non-hits. Selected curves were within 1% of the mean AUC value of 1,000 training/validation trials.
(D) Heatmaps showing AUC values for individual epigenomic features for classifying coding and lncRNA hits versus non-hits. Statistical significance determined at the 99% confidence level from 1,000 bootstraps; non-significant features denoted in gray.
(E) ChIP-seq profiles showing average H3K4me3 signal in a 2-kb window at the promoter region of coding and lncRNA genes in ESCs. Coding hits in green, lncRNA hits in magenta, and non-hits in gray.
(F) Odds ratio for the enrichment of hits in broad H3K4me3 domains. Both coding and lncRNA gene hits were significantly enriched compared with non-hits. Dashed line denotes an odds ratio of 1 (null hypothesis), and error bars denote 95% confidence intervals by Fisher exact test. ∗p < 1 × 10−8.
The epigenomic data classified both coding and lncRNA gene hits at a similar performance, with mean AUC of 0.75 and 0.74, respectively (Figure 4C). To explore this finding at a more granular level, we turned to the analysis of individual histone marks (Figure 4D), which revealed significant scores for histone-3 lysine-4 trimethylation (H3K4me3), indicating that this mark distinguished hits from non-hits better than random chance for both coding and lncRNA genes. The H3K4me3 modification is associated with active genes,30 with the top 5% “broadest” domains enriched for genes important for cellular identity and function.31 Analysis of chromatin immunoprecipitation sequencing (ChIP-seq) profiles revealed elevated H3K4me3 deposition at both coding and lncRNA hit promoter regions (Figure 4E). Additionally, both coding and lncRNA hits were significantly enriched (odds ratio, ∼4–8) in the broadest H3K4me3 domains (Figure 4F), indicating importance in cell identity. Together, these findings illustrate how epigenomic features—as a group as well as at the level of a specific histone mark—distinguish hits from non-hits in a screen for regulators of neural induction.
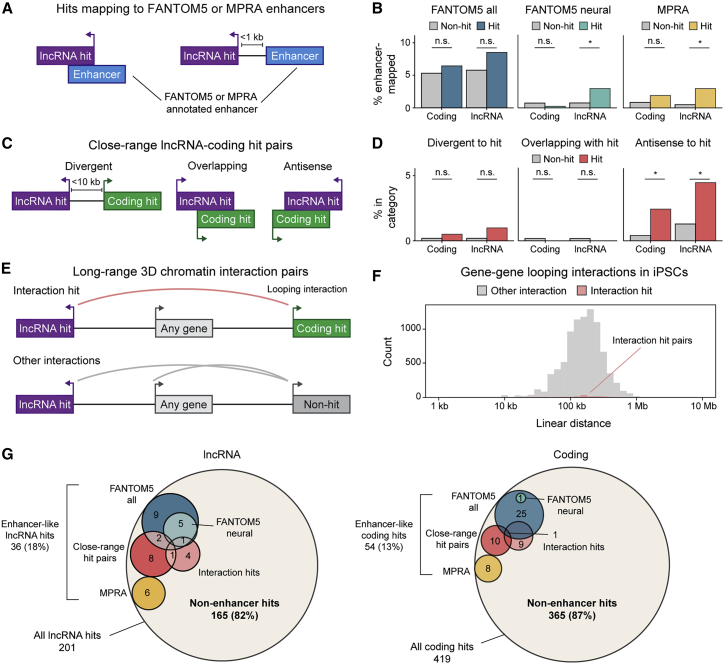
A small fraction of lncRNA gene hits have evidence of enhancer-like function
Some lncRNA loci can function as transcriptional enhancers.32,33,34 We therefore investigated what proportion of lncRNA hits have evidence of enhancer-like function. The linear genomic distance of lncRNA gene hits to coding gene hits was somewhat decreased (median, 1.4 Mb) compared with the overall distribution (2 Mb), although these distributions were largely overlapping (Figure S5A). To more comprehensively identify potential enhancer loci among hits, we considered the Functional ANnoTation Of the Mammalian genome (FANTOM5) atlas of 43,011 human enhancers,35 a massively parallel reporter assay (MPRA) that identified 1,547 candidate regulatory sequences activated during human neural induction,36 the genomic relationship of each lncRNA gene hit with the nearest coding gene hit, and long-range three-dimensional intrachromosomal interactions between lncRNA and coding genes derived from Proximity ligation-assisted ChIP-seq (PLAC-seq) (Figures 5A–5F). In total, 18% (36 of 201) of lncRNA hits overlapped at least one of these maps (Figure 5G, left). Of note, these broadly inclusive criteria also classified 13% (54 of 419) of coding hits as potential enhancers (Figure 5G, right). At higher stringency—evidence from at least two of the analyses—only 2% (4 of 201) of lncRNA hits were classified as enhancers (Tables S3 and S4). Thus, only a minority of coding and lncRNA gene hits are potential enhancers.
Figure 5.
A small fraction of lncRNA gene hits have evidence of enhancer-like function
(A) Diagram of screen hits that map to enhancer sequences across FANTOM5 atlas and MPRA neural induction enhancer datasets.
(B) Bar plots showing fraction of coding and lncRNA genes (colored, hit; gray, non-hit) mapping to enhancers described in (A).∗p < 0.05 comparing hits and non-hits by Fisher exact test; n.s., non-significant.
(C) Diagram of close-range lncRNA-coding hit pair genomic relationships.
(D) Bar plots showing fraction of coding and lncRNA genes (colored, hit; gray, non-hit) in each category described in (C). ∗p < 0.05 comparing hits and non-hits by Fisher exact test; n.s., non-significant.
(E) Diagram of long-range 3D chromatin interactions between lncRNA and coding hit pairs.
(F) Histogram showing distances of long-range 3D gene-gene looping interactions identified by PLAC-seq. Interaction hits are colored in light red; all other interactions in gray.
(G) Venn diagrams showing potential enhancer-like screen hits classified into the categories detailed above.
Dual genome-wide screens enable Perturb-seq experiment to dissect coding and lncRNA phenotypes
By coupling CRISPRi genetic perturbation with rich single-cell transcriptomic readout, Perturb-seq16,17,18 provides deeper insights into gene function and cell biology. While the readout of pooled screens is usually based on simple phenotypes such as cell growth, survival, or marker gene expression, Perturb-seq allows the dissection of different phenotypes and molecular mechanisms that are masked in bulk experiments.
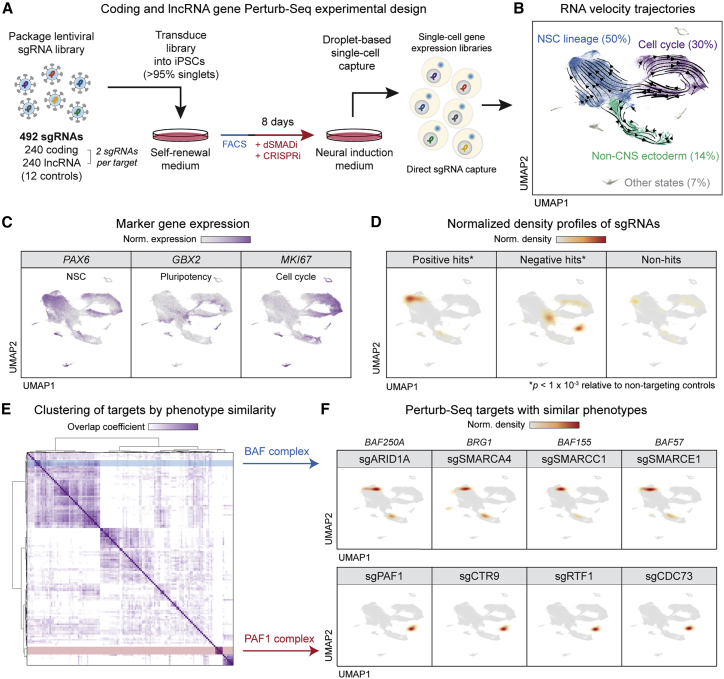
We used our functional atlas from the dual genome-wide screens to inform a Perturb-seq experiment that interrogates both coding and lncRNA gene function. We selected targets by prioritizing the highest scoring differentiation hits and excluding any hits with strong proliferative phenotypes; i.e., those with absolute γ greater than 1 (predicted to become substantially overrepresented or underrepresented due to survival differences). For comparative analysis, we also randomly sampled non-hit genes with similar expression levels. The final Perturb-seq library consisted of 480 sgRNAs for 240 unique targets (120 lncRNA and 120 coding genes, with two independent sgRNAs for each target), covering 60 positive differentiation hits, 85 negative differentiation hits, 30 dual hits, and 65 non-hits; additionally, 12 non-targeting control sgRNAs were included for a total of 492 unique sgRNAs. The library was transduced into CRISPRi-iPSCs at a low multiplicity of infection (MOI) of 0.1, corresponding to >95% cells with a single sgRNA integration. After FACS for sgRNA+ cells, we initiated neural induction and activation of CRISPRi (Figure 6A). On day 8, we harvested cells and prepared single-cell RNA-seq (scRNA-seq) libraries using direct sgRNA capture.18
Figure 6.
Dual genome-wide screens enable Perturb-seq experiment to dissect coding and lncRNA phenotypes
(A) Overview of neural induction Perturb-seq experimental design with direct capture of sgRNAs.
(B) Major trajectories derived from RNA velocity and visualized by UMAP.
(C) Expression of markers for NSCs, pluripotent cells, and cycling cells, visualized on UMAP.
(D) Normalized density heatmaps of hit and non-hit sgRNAs on UMAP. Density profiles of target sgRNAs were calculated in UMAP space and normalized to the background density of non-targeting controls. ∗p < 1 × 10−3 based on multivariate Kolmogorov-Smirnov test versus non-targeting control distribution.
(E) Heatmap of hierarchical clustering of Perturb-seq targets by similarity of sgRNA density profiles by overlap coefficient. Two groups of targets, the BAF and PAF1 complexes, are highlighted in blue and red, respectively.
(F) Normalized density heatmaps of sgRNAs targeting BAF and PAF1 complex members, showing their colocalization in UMAP.
Following sequencing and data processing, we filtered cells for sgRNA detection, singlet status, and quality metrics (STAR Methods; Table S5). We obtained a total of 78,393 cells that harbored single sgRNA perturbations, with each perturbation represented in a median of 317 cells. Analysis of target gene expression data revealed a median knockdown efficiency of 80% (Figure S6A), comparable with prior studies.18 The Perturb-seq dataset was visualized in two dimensions using uniform manifold approximation and projection (UMAP).
Based on RNA velocity analysis37,38 (Figure 6B) and marker gene expression (Figures 6C and S6B), we identified three major cellular trajectories. The largest trajectory (NSC lineage, representing ∼50% of the cells) corresponded to non-cycling cells undergoing neural induction, with velocities directed toward a final cell state with high expression of neural markers including PAX6, FOXG1, and EMX2. Pluripotency markers such as GBX2 and POU5F1/OCT4 were lowly expressed in this trajectory but present in other cell populations (Figures 6C and S6B). The second largest trajectory (cell cycle, ∼30% of cells) consisted of actively cycling cells (CDC6+ and MKI67+), including both PAX6+ cells as well as PAX6− cells that expressed pluripotency markers. Cells exiting the cell cycle trajectory branched into either the NSC lineage or a third trajectory—non-central nervous system (non-CNS) ectoderm, ∼14% of cells—characterized by markers of ectodermal lineages (e.g., TFAP2A/B) that normally develop outside the CNS and can appear in a subset of cells undergoing neural induction.21 Approximately 7% of cells did not fall within these three major trajectories.
Each Perturb-seq sgRNA was mapped to each cell, and we constructed normalized 2D density heatmaps to visualize the enrichment of hits and non-hits in the UMAP space. As a group, positive-hit sgRNAs were enriched in PAX6+ NSCs, whereas negative-hit sgRNAs were enriched in multiple PAX6− cell states (Figure 6D). The group of non-hit sgRNAs were not statistically distinguishable from non-targeting control sgRNAs (representing non-perturbed cells), indicating that they did not have substantial effects on the neural induction transcriptome. Thus, Perturb-seq validates the differentiation phenotypes of targets from the genome-wide screens.
To assess potential sgRNA effects on cell proliferation, we quantified the number of cells expressing each sgRNA, providing a relative measure of this growth phenotype (e.g., a target that reduces proliferation would drop out over time, resulting in fewer sgRNA+ cells). For targets in the Perturb-seq experiment, the sgRNA cell counts at day 8 of neural induction were proportional to the γ proliferation phenotype from the genome-wide screens (Figure S6C), with dual hits showing a strong correlation (Pearson r = 0.92). Thus, Perturb-seq confirms both proliferation and differentiation phenotypes, supporting the findings of the genome-wide screens (Figures 3C and 3D).
To study the effects of individual hits, we generated normalized density heatmaps for each target, using density-based spatial clustering and application with noise39 (DBSCAN) to identify the discrete UMAP regions of high sgRNA density (STAR Methods). Pairwise analysis and clustering of the sgRNA density profiles revealed groups of targets that had similar effects (Figures 6E, S6D, and S6E; Table S6). For instance, BAF1 complex members were positive hits in the genome-wide screen (Figures 1E and S2B), and sgRNAs targeting ARID1A (BAF250A), SMARCA4 (BRG1), SMARCC1 (BAF155), and SMARCE1 (BAF57) were localized in the same patterns in UMAP (Figure 6F, top), suggesting they affected neural induction in a similar manner. Knockdown of the BAF complex led to cells farther along both NSC and non-CNS ectoderm trajectories, consistent with the role of this chromatin regulator complex in maintaining pluripotency and acting as a general barrier to differentiation.40 Proteins encoded by negative hits PAF1, CTR9, RTF1, and CDC73 physically interact in a complex known as PAF1c that regulates transcription, chromatin structure, and signaling pathways important for embryogenesis.41 Targeting these PAF1c components produced a transcriptome that is distinct from the major cell trajectories observed in neural induction (Figure 6F, bottom). Similarly, Perturb-seq revealed overlapping phenotypes among physically interacting hits related to the Mediator, DNA synthesis, and Polycomb complexes (Figure S6D).
Additionally, genes that function in the same pathway produced similar UMAP density profiles. POU5F1/OCT4 is upregulated by SALL4,42 and the density heatmaps of these two positive hits were highly overlapping (Figure S6E, left), indicating that repression of POU5F1 and SALL4 led to similar phenotypes. Analysis of the density heatmaps also identified similar patterns among other coding genes that function in the same pathways, such as Wingless (WNT) and mitogen-activated protein kinase (MAPK) signaling (Figure S6E). Collectively, these examples demonstrate that Perturb-seq targeting of genes in the same pathway or molecular complex produces highly similar UMAP profiles that reflect the underlying biological process governed by those genes.
Coding gene repression stalls or aborts differentiation, while lncRNA gene repression permits a greater diversity of cell states
Based on the analysis of density profiles for all Perturb-seq targets, we identified a total of 29 cell states (Figures 7A and S7A–S7C). Each Perturb-seq target was then analyzed for the relative distribution of its sgRNAs mapping to each of the 29 states, and these data were visualized by heatmap (Figure S7D). Hits were color coded according to their positive or negative differentiation phenotype from the dual genome-wide screens, and, although this information was not used to inform clustering, positive and negative hits segregated from each other. For instance, positive hits associated with NSC states (e.g., 16, 12, 9, 23, 24), while negative hits were prominent in less differentiated, intermediate cell states (e.g., 13, 6).
Figure 7.
Coding gene repression stalls or aborts differentiation, while lncRNA gene repression permits a greater diversity of cell states
(A) Cell states associated with target perturbations, on UMAP.
(B) Normalized density heatmaps of positive and negative coding and lncRNA hits with similar magnitudes of effect from the genome-wide screens.
(C) Heatmap of gene expression signatures of cell states observed in (B), with examples of enriched ontology terms (FDR < 0.01).
(D) Pie charts showing the proportion of negative hits classified as non-productive or multiple trajectories. Coding and lncRNA hits were significantly different in the proportion of these phenotypes. ∗p < 1 × 10−5 by Fisher’s exact test.
(E) Diagram of internally controlled differentiation assay, with sgRNA-containing cells labeled by GFP or RFP.
(F) Overall percentage of GFP and RFP cells that express TAGLN protein (n = 3 replicates per condition). GFP/RFP double-positive cells (expressing both sgRNAs) were excluded. Error bars denote 1 SD. ∗p < 0.01 by t test.
(G) Relative expression of SERTAD4-AS1 and SERTAD4 coding gene in Perturb-seq experiment in cells with sgSERTAD4-AS1 compared with sgControl. Compared with control cells, complete knockdown was achieved for SERTAD4-AS1, with no significant change in SERTAD4 coding gene expression. Error bars show mean ± SEM (n = 6 pseudobulk samples). ∗p = 0.004 by t test.
For both coding and lncRNA genes, positive hits generally produced similar PAX6+ NSC states. For instance, sgRNAs targeting OGT—which encodes the O-GlcNAc transferase protein that regulates pluripotency and neural differentiation43,44—were enriched in NSC state 16 (Figure 7B). This state was characterized by the highest expression of neural markers, including genes involved in forebrain development (e.g., PAX6, FOXG1, FEZF1, EMX2) (Figure 7C; Table S6). Targeting the novel lncRNA gene LH09400 (internal identifier) led to enrichment in NSC state 12 (Figure 7B), which expressed a highly similar signature of neural markers but with elevated levels of HES4, HES5, and ID4, genes downstream of NOTCH signaling (Figure 7C).
Negative hits showed highly divergent phenotypes between coding genes and lncRNA genes. For coding genes, the most common phenotype (40%) was enrichment in an intermediate cell state. For example, knockdown of the forebrain development factor HESX1, a homeobox,45 led to enrichment in cell state 6. This intermediate state lies at the junction of the major RNA velocity trajectories (Figure 6B) and is characterized by PAX6−/GBX2+ cells exiting the cell cycle (Figure 6C), suggesting that these cells are most similar to undifferentiated cells, and may have stalled or are slower to progress along their differentiation trajectory to NSCs. The next most common phenotype (16%) was an apoptotic signature (e.g., BAX, CDKN1A), indicating that these cells failed differentiation, most likely due to impaired survival. Together, stalled and apoptotic phenotypes (collectively “non-productive”) represented the majority (56%) of negative coding hits.
In contrast, few lncRNA gene perturbations exhibited non-productive states as the main phenotype. Repression of lncRNA genes instead generally led to diverse cell states along multiple trajectories (Figure S7E). For instance, sgRNAs targeting the uncharacterized lncRNA gene SERTAD4-AS1 were enriched in all three trajectories (Figure 7B), even though this hit inhibited neural induction to a similar degree as HESX1. In addition to affecting cells in the NSC lineage (state 12), perturbation of SERTAD4-AS1 also resulted in changes to cell cycle (state 18), and cells at the far end of the non-CNS ectoderm trajectory that appear neural crest derived (state 1; Figure 7C). Thus, despite inhibiting neural induction to a similar degree, the underlying phenotype of SERTAD4-AS1 was vastly different from that of HESX1.
Quantitative classification of negative hit phenotypes revealed profound differences between coding and lncRNA genes (Figure 7D). Although coding gene knockdown typically prevented neural induction by generating non-productive (i.e., stalled or apoptotic) phenotypes, lncRNA gene knockdown generally blocked neural induction by dispersing cells along multiple trajectories, including cell identities outside the NSC lineage. Furthermore, the number of trajectories was not explained by neural induction effect size (Figure S7F). These granular Perturb-seq phenotypes therefore support our broader findings of differences in coding and lncRNA differentiation and proliferation phenotypes (Figures 3D and 3E). Collectively, our findings indicate that coding and noncoding genes required for neural induction have markedly different phenotypes, suggesting that lncRNA genes—which have arisen much later in evolution than coding genes—may be employed by the genome for broadly different cellular roles, providing an additional facet of complex gene regulation during development.
Repression of SERTAD4-AS1 increases production of TAGLN+ cells
To facilitate the widespread use of our resource, we created an interactive data portal (danlimlab.shinyapps.io/dualgenomewide). This website enables intuitive exploration of our collective datasets without any programming experience, from retrieving the differentiation and proliferation effects for genes of interest to visualizing the single-cell gene expression and sgRNA density profiles from the Perturb-seq experiment (Figure S8A).
For instance, Perturb-seq revealed that targeting the SERTAD4-AS1 gene inhibited neural induction by causing a multiple trajectory phenotype, producing cells with transcriptomes far outside of the neural stem cell lineage, such as non-CNS neural crest. The SERTAD4-AS1 gene is located on chromosome 1 and produces multiple multi-exon isoforms, in antisense orientation to a transcript isoform of the coding gene SERTAD4. From the transcriptomics data, SERTAD4-AS1 expression is highest in iPSCs and induced cells (TPM > 1), with decreased expression in the transition between these two states (Figure S8B). Importantly, the dual genome-wide nature of the study enabled us to determine that, while the SERTAD4-AS1 lncRNA gene was a differentiation hit, SERTAD4 coding gene was a non-hit (Figure S8C). In our enhancer analysis, SERTAD4-AS1 did not map to any MPRA or FANTOM5 enhancers, and chromatin interaction analysis did not find any statistically enriched long-range 3D interactions with other hits. Using the data portal to explore the Perturb-seq analysis for SERTAD4-AS1 revealed the transgelin (TAGLN) gene—which encodes an actin-binding protein and early marker of smooth muscle cell differentiation46,47—to be the top marker associated with SERTAD4-AS1 perturbation (Figure S8D).
To further explore the underlying biological phenotype of SERTAD4-AS1 gene repression, we designed an internally controlled differentiation assay. We sparsely labeled CRISPRi-iPSCs with viral sgRNA vectors targeting SERTAD4-AS1 (co-expressing GFP) or a non-targeting control sequence (co-expressing RFP) (Figure 7E). After 8 days of neural induction, we performed immunofluorescent staining for TAGLN protein. Significantly more GFP+ cells (sgSERTAD4-AS1) were positive for TAGLN protein compared with RFP+ cells (sgControl), indicating that loss of SERTAD4-AS1 during neural induction leads to the abnormal generation of cells with this early marker of smooth muscle (Figures 7F and S8E). Notably, analysis of cells containing sgSERTAD4-AS1 in the Perturb-seq experiment revealed excellent on-target knockdown of SERTAD4-AS1 without affecting SERTAD4 coding gene expression (Figure 7G). These experimental findings, as predicted by our genome-wide screens and Perturb-seq analyses, further strengthen the neural induction phenotype of this lncRNA gene hit.
Discussion
In addition to identifying hundreds of coding and lncRNA genes that regulate neural induction, the scale of the dual genome-wide screens provided fundamental insights that would not have been apparent with less comprehensive approaches. Perturb-seq additionally revealed surprising differences in the phenotypes of coding and lncRNA genes when examined at high resolution. Taken together, our systematic studies underscore the unique functional roles of the lncRNA and coding genomes and have important implications for our understanding of gene expression studies, genome evolution, and developmental phenotypes.
Gene expression is often used to predict biological function in development.7,48 In our systematic analyses, the inference of function by transcriptional information was relatively strong for coding genes, but much weaker for the lncRNA class. In contrast, epigenomic information (e.g., the level of specific histone modifications) distinguished hits from non-hits for both classes. Only a minority of coding and lncRNA hits mapped to potential enhancers, suggesting that most hits do not regulate neural induction through such activity. In addition to providing information that can help prioritize lncRNA genes for functional studies, these insights broadly influence the interpretation of expression data in other biological contexts and certain disease-association studies, highlighting the critical need for functional data rather than reliance on descriptive data (e.g., expression patterns).
Coding genes were equally distributed between positive and negative regulators of neural induction, whereas lncRNA genes were strongly enriched for positive regulators. Remarkably, analysis of growth effects uncovered further differences between the two classes: lncRNA genes were ∼10-fold enriched for roles in differentiation, whereas coding genes preferentially regulated proliferation. Given their tissue-specific expression and recent expansion in evolution, lncRNA genes have been suggested to play critical developmental roles, especially in the mammalian nervous system.2,7,8,9,10,49,50 Our work provides systematic, genome-wide functional evidence that the lncRNA class is enriched for specialized cellular roles (e.g., regulating differentiation) rather than essential housekeeping roles,51,52,53 which are dominated by protein-coding genes.
The genome-wide screen resource enables highly granular experimental studies, such as our coding-lncRNA gene Perturb-seq experiment. By targeting hundreds of coding and lncRNA genes identified as functional in neural induction and studying transcriptomes at single-cell resolution, we dissected these phenotypes in new detail. Remarkably, most negative coding hits stalled or aborted the NSC trajectory upon knockdown, whereas lncRNA gene knockdown was more permissive of diverse states, including those outside of the NSC lineage. For example, knockdown of HESX1 produced a single, intermediate PAX6− state in the NSC trajectory (Figure 7B), suggesting that these cells become stalled in their differentiation. In contrast, knockdown of lncRNA SERTAD4-AS1—a hit with a similar overall phenotype magnitude as HESX1—was enriched in multiple diverse states, including cell types outside the NSC lineage, such as neural crest cells with early markers of smooth muscle.
One interpretation of this comparison is that, in neural induction, HESX1 may function primarily along a specific developmental program, whereas lncRNA SERTAD4-AS1 has function(s) dispersed across multiple cellular programs; for example, immunofluorescence staining revealed that repression of SERTAD4-AS1 produced significantly more cells positive for the TAGLN protein, a canonical marker of smooth muscle cells, suggesting that SERTAD4-AS1 promotes neural induction by suppressing other developmental programs. More generally, the collective results from this Perturb-seq study suggest a conceptual model in which lncRNA hits are enriched for function in “shepherding” cells through the differentiation process, helping prevent cells from “escaping” into non-intended cell trajectories. It is unclear whether this phenotypic heterogeneity associated with lncRNA genes is due to modifying factors, stochasticity, or other unmeasured processes, and detailed analyses of individual genes will be necessary to elucidate the underlying mechanisms. Regardless, these findings emphasize the distinct roles played by the coding and lncRNA genomes in human cell differentiation.
Our systematic functional studies showcase the fundamental and surprising differences between the coding and lncRNA genomes, and provide an expansive dual genome-wide resource for investigating their function in human cell differentiation. A variety of studies have implicated both protein-coding and lncRNA genes in a wide range of neurodevelopmental disorders.48,54,55,56,57 Our work reveals that the lncRNA genome enables proper differentiation in critical and unexpectedly unique ways from the coding genome, providing a functional context in which to begin studying potential disease associations. More generally, this vast trove of functional data across the human genetic landscape enables fundamental biological insights that are difficult to obtain by individual gene studies or even screens of the coding or lncRNA genome alone.
Limitations of the study
The conclusions of our study are limited by the use of iPSCs rather than ESCs. The epigenetic memory of iPSCs may predispose them to specific differentiation pathways related to the parental cell type from which they were derived. Studies in other human pluripotent stem cells and in other developmental contexts (e.g., mesodermal lineages) will be necessary to generalize the findings from this work. Additionally, our characterization of SERTAD4-AS1 in this study has been limited. Full analysis of a lncRNA gene cannot rely on CRISPRi alone and requires many detailed mechanistic studies, such as the case for lincRNA-p21.58,59,60 Although we did not find that knockdown of SERTAD4-AS1 affected the expression of the neighbor coding gene, its targets and molecular mechanism are unknown. As CRISPRi can perturb a broad range of function—from cis-regulatory activity to RNA-dependent trans function—genome engineering strategies such as promoter deletion or poly(A) terminator insertion32,61 will be particularly important for understanding the function of this gene.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-PAX6 | BD Biosciences | Cat# 561462, RRID: AB_10715442 |
| Rabbit anti-TAGLN | Abcam | Cat# ab14106, RRID: AB_443021 |
| Chicken anti-GFP | Aves Labs | Cat# GFP-1020, RRID: AB_10000240 |
| Goat anti-RFP/tdTomato | SICGEN | Cat# AB8181, RRID: AB_2722750 |
| Donkey anti-Rabbit Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573, RRID: AB_2536183 |
| Donkey anti-Goat Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21432, RRID: AB_2535853 |
| Donkey anti-Chicken Alexa Fluor 488 | Jackson ImmunoLabs | Cat# 703-545-155, RRID: AB_2340375 |
| Bacterial and virus strains | ||
| MegaX competent Cells | Thermo Fisher Scientific | Cat# C640003 |
| Chemicals, peptides, and recombinant proteins | ||
| SB431542 | Selleckchem | Cat# S1067 |
| LDN193189 | Selleckchem | Cat# S2618 |
| Y-27632 ROCK inhibitor | Selleckchem | Cat# S1049 |
| Doxycycline | Selleckchem | Cat# S4163 |
| Puromycin | Tocris | Cat# 408950 |
| Essential 8 (E8) medium | Thermo Fisher Scientific | Cat# A1517001 |
| Essential 6 (E6) medium | Thermo Fisher Scientific | Cat# A1516401 |
| StemPro Accutase | Thermo Fisher Scientific | Cat# A1110501 |
| TransIT-LT1 | Mirus Bio | Cat# MIR 2300 |
| ViralBoost | Alstem Bio | Cat# VB100 |
| Q5 High-Fidelity Master Mix | NEB | Cat# M0492 |
| Critical commercial assays | ||
| Direct-zol RNA Miniprep | Zymo Research | Cat# R2050 |
| NEBNext Ultra II Directional RNA Library Prep Kit | NEB | Cat# E7760 |
| NEBNext rRNA Depletion Kit | NEB | Cat# E6350 |
| DNAStorm FFPE DNA Extraction Kit | CellData | Cat# CD502 |
| Chromium Single-Cell 3′ v3 with Feature Barcoding | 10x Genomics | Cat# PN-1000075, PN-1000153, PN-1000079 |
| High Sensitivity RNA ScreenTape | Agilent | Cat# 5067-5580 |
| High Sensitivity D1000 DNA ScreenTape | Agilent | Cat# 5067-5584 |
| High Sensitivity D5000 DNA ScreenTape | Agilent | Cat# 5067-5592 |
| Deposited data | ||
| Raw and analyzed data | This study | GEO: GSE150062 |
| ENCODE Roadmap Epigenomics raw data | Xie et al.29 | GEO: GSE16256 |
| FANTOM5 human enhancer atlas | Andersson et al.35 | https://fantom.gsc.riken.jp/5/ |
| MPRA neural induction enhancers dataset | Inoue et al.36 | GEO: GSE115046 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Experimental models: Cell lines | ||
| CRISPRi WTC11 iPSCs | Mandegar et al.62 | Gen1C |
| Lenti-X 293T | Takara Bio | Cat# 632180 |
| Oligonucleotides | ||
| Perturb-seq sgRNA sequences | Twist Biosciences | Table S5 |
| Recombinant DNA | ||
| pCRISPRia-v2 sgRNA vector | Horlbeck et al.22 | Cat# 84832 |
| pBA904 Perturb-seq sgRNA vector | Replogle et al.18 | Cat# 122238 |
| Software and algorithms | ||
| ScreenProcessing | Gilbert et al.63 | https://github.com/mhorlbeck/ScreenProcessing |
| Perturb-seq sgRNA assignment | Replogle et al.18 | https://github.com/josephreplogle/guide_calling |
| MAPS pipeline | Juric et al.64 | https://github.com/ijuric/MAPS |
| Original code for analyses | This study | https://github.com/symbiologist/dualgenomewide; https://doi.org/10.5281/zenodo.6815996 |
| ENCODE ChIP-Seq pipeline | ENCODE Project Consortium | https://github.com/ENCODE-DCC/chip-seq-pipeline2 |
| CellRanger 4.0.0 | 10x Genomics | http://software.10xgenomics.com/ |
| HISAT2 | Pertea et al.65 | http://daehwankimlab.github.io/hisat2/ |
| kallisto 0.45.0 | Bray et al.66 | https://pachterlab.github.io/kallisto/about |
| FastQC 0.11.8 | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| featureCounts 2.0.0 | Liao et al.67 | http://subread.sourceforge.net/ |
| gffcompare 0.10.6 | Pertea et al.68 | https://ccb.jhu.edu/software/stringtie/gffcompare.shtml |
| bbduk 38.36 | DOE Joint Genome Institute | https://sourceforge.net/projects/bbmap/ |
| tximport 1.12.3 | Soneson et al.69 | https://github.com/mikelove/tximport |
| DESeq2 1.24.0 | Love et al.70 | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| maSigPro 1.56.0 | Conesa et al.71 | https://www.bioconductor.org/packages/release/bioc/html/maSigPro.html |
| caret 6.0 | Kuhn72 | https://topepo.github.io/caret/ |
| Seurat 3.9.0 | Stuart et al.73 | https://satijalab.org/seurat/ |
| velocyto | Manno et al.37 | http://velocyto.org/ |
| scVelo | Bergen et al.38 | https://scvelo.readthedocs.io/ |
| MACS2 2.2.6 | Zhang et al.74 | https://github.com/macs3-project/MACS |
| deepTools 3.4.0 | Ramirez et al.75 | https://deeptools.readthedocs.io |
| Other | ||
| Interactive data resource | This study | https://danlimlab.shinyapps.io/dualgenomewide |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daniel Lim (daniel.lim@ucsf.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell lines
The male CRISPRi wild-type C human induced pluripotent stem cell line62 was obtained from and verified by the Gladstone Institutes Stem Cell Core. Cells were maintained in Essential 8 medium (Thermo Fisher Scientific) on Matrigel (Corning).
Method details
Neural induction of human pluripotent stem cells
Neural induction of human iPSCs was performed using the dual SMAD inhibition paradigm.6,21 Engineered CRISPRi-iPSCs62 were grown in Essential 8 (Thermo Fisher Scientific) media on Matrigel (Corning) to 80% confluency. Cells were rinsed with DPBS and dissociated with Accutase (StemPro). After centrifugation at 300 x g for 3 min and resuspension in Essential 8 media with 10 μM Y-27632 ROCK inhibitor (Selleckchem), cells were replated at a density of 250,000 cells/cm2 overnight at 37°C. The next day (D0), cells were rinsed with DPBS and changed to neural induction media, which consisted of Essential 6 media (Thermo Fisher Scientific) with freshly-added SMAD inhibitors 500 nM LDN193189 (Selleckchem) and 10 μM SB431542 (Selleckchem). Media was replaced every 2 days until the endpoint of interest, as described.21
Flow cytometry and fluorescence-activated Cell sorting (FACS)
Cells were harvested by dissociation with Accutase. After washing twice with DPBS, cells were quantified on the Countess II (Thermo Fisher Scientific) and resuspended in 4% paraformaldehyde at 10 million cells/ml for 20 min at room temperature. Cells were then washed twice in a permeabilization buffer (DPBS 5% goat serum with 0.5% saponin) and blocked in the same buffer for 30 min at room temperature. Primary and secondary antibodies were added for 30 min each with 2 washes in between. Prior to running through the instrument, cells were resuspended in DPBS 5% BSA at 5 million cells/ml. Cells were gated by size (FSC) and granularity (SSC) and then for singlets by FSC-H vs. FSC-W followed by SSC-H vs. SSC-W. Cells were further gated for expression of dCas9-KRAB by coexpression of mCherry and expression of sgRNAs by coexpression of BFP before analyzing stained proteins.
Immunocytochemistry
Cells were grown on glass chamber slides until the desired endpoint and fixed in 4% paraformaldehyde for 15 min. After two washes in PBS, cells were permeabilized in 0.1% Triton X-100 for 15 min, followed by blocking with 10% goat serum for 1 h. Primary and secondary antibodies were added for 60 min each with 2 washes in between. DAPI (Thermo Fisher Scientific) was added 1:1,000 with the species-specific secondary antibody (see Key resources table). Slides were mounted overnight with coverslips using Aqua Poly/Mount (Polysciences).
RNA purification and sequencing library preparation
RNA was purified using Direct-zol (Zymo) columns with DNase I treatment. RNA integrity was verified using the TapeStation 4200 (Agilent) prior to library generation. Libraries were generated using the NEBNext Ultra II Directional RNA kit (NEB) according to the manufacturer’s instructions. For polyA RNA, Oligo-dT magnetic bead selection was used prior to library generation. For total RNA, rRNA depletion using hybridization and RNase H-induced cleavage was used prior to library generation. For all samples, 2 μg of RNA was used as input. Samples were sequenced on HiSeq 4000 to >160 M reads per time-point.
Genome-wide CRISPRi screens for neural induction
Sublibraries of the hCRISPRiv2 and CRiNCL sgRNA libraries were assembled into coding and lncRNA libraries at equimolar ratios and sequenced to ensure uniform distribution. Coding and lncRNA screens were performed separately on a staggered schedule for feasibility. A lncRNA sublibrary (common sublibrary) was also performed in a separate batch to validate proliferation phenotypes (described below in “Differentiation and proliferation screen analysis”) and results were aggregated with the main lncRNA library during analysis. Lentivirus was prepared in the high titer Lenti-X 293T subclonal line (Clontech) using TransIT-LT1 (Mirus) and ViralBoost (Alstem) according to the manufacturer’s instructions. Viral supernatant was collected at 72 h, filtered, and concentrated through ultracentrifugation for 2 h at 4°C. Titer was assessed through serial dilution of single-freeze aliquots to determine the necessary amount of virus to achieve the desired MOI of 0.5. In total, approximately 650 million CRISPRi-iPSCs were transduced for coding and lncRNA libraries (each with 2 independent replicates). Once transduced and seeded, replicates were maintained independently and never re-pooled. Transduced iPSCs were selected under puromycin to >80% positivity and allowed 2 days to recover with >1,000X sgRNA library representation per replicate. At the next seeding, an additional aliquot of each sample (∼100 M cells per replicate) was frozen to measure the initial sgRNA abundance (“T0”). A second, small aliquot of cells was seeded in a sentinel 6W plate for monitoring screen progress. Doxycycline was added at 1 μM at the initiation of neural induction to activate the dCas9-KRAB CRISPRi machinery. At days 6–8, cells from the sentinel plate were fixed, permeabilized, and stained to assess neural induction progress. All samples were harvested, fixed, permeabilized, stained, and sorted into PAX6+ and PAX6- fractions (top and bottom thirds) as described above with final coverage of ∼4000X at day 8. Selection of this time-point was based on the flow cytometry analysis (Figure 1B) showing presence of both PAX6+ and PAX6- populations; earlier time-points may not allow for sufficient differentiation and would enable the discovery of sgRNAs that accelerate neural induction, but not those that prevented it. Later time-points, on the other hand, may preclude the enrichment of sgRNAs that promoted neural induction, and would mainly identify depleted sgRNAs. Genomic DNA was harvested using a chemically-catalyzed FFPE extraction method (CellData) following the manufacturer’s instructions. Sequencing libraries were prepared by targeted amplification of integrated sgRNAs using Q5 High-Fidelity Master Mix (NEB) and sequenced on an Illumina HiSeq 4000 to >50 M reads per replicate. Processing of screen data was performed as previously described using ScreenProcessing.63
Individual sgRNA cloning and experiments
Individual sgRNAs were cloned using an annealing and ligation procedure, as previously described.63 Sense and antisense oligos that matched the desired CRISPRi protospacer sequence were annealed and ligated into a U6-driven lentiviral expression vector derived from pSico. All individually-cloned sgRNA vectors underwent Sanger sequencing to verify successful sequence insertion. Lentivirus was prepared and transduced into CRISPRi-iPSCs as described above, except in an arrayed fashion (12-well plates). Transduction efficiency was monitored through detection of a BFP cassette coexpressed by the sgRNA vector. For validation experiments, neural induction was performed as described above and cells were harvested at the endpoint for flow cytometry. Assessment of individual sgRNA phenotypes were performed by comparing the ratio of PAX6+ to PAX6- cells for populations with and without sgRNA (measured by BFP). For immunocytochemistry experiments, GFP and RFP versions of the same vectors were used and cells were plated on glass slides and analyzed as described above (“Immunocytochemistry”).
Proximity ligation-Assisted ChIP-Seq (PLAC-Seq) for long-range chromatin loops
PLAC-Seq was performed as previously described.76,77 Approximately 1–5 million cells were used for library preparation. Digestion was performed using 100 U MboI for 2 h at 37°C, and chromatin immunoprecipitation was performed using Dynabeads M-280 sheep anti-rabbit IgG (Invitrogen 11203D) superparamagnetic beads bound with 5 μg anti-H3K4me3 antibody (Millipore 04-745). Sequencing adapters were added during PCR amplification. Libraries were sequenced at paired-end 150 on an Illumina HiSeq 4000. Quality and adaptor trimming were performed with fastp (0.13).
Perturb-seq experimental design
A direct-capture Perturb-seq library consisting of a mixture of 492 sgRNAs targeting 120 coding genes, 120 lncRNAs, and 12 non-targeting control sequences was cloned as a pool as previously described.18 Targets were selected from those with the highest genome-wide neural induction screen scores, with strong proliferation hits excluded due to expected growth dropout effects. Dual hits with mild-moderate phenotypes (γ between −1 and 1) were not excluded from the experiment. A set of lncRNA sgRNAs identified as targeting ambiguous loci (described below, under “Differentiation and proliferation screen analysis”) were included in the Perturb-seq library for assessing potential local effects, and these targets were excluded for all reported analysis. Reported analyses included the remaining 195 targets. After library sequencing to ensure sgRNA uniformity, CRISPRi-iPSCs were transduced with the Perturb-seq library at a low MOI of 0.1 (corresponding to >95% of cells with a single sgRNA integration) and >1,000X coverage using the lentiviral protocol described above. After FACS to sort for sgRNA + cells, we recovered the cells in maintenance media for 2 days before initiating dual SMAD inhibition neural induction. We performed single-cell and direct sgRNA capture at our previously determined endpoint of day 8, aiming for >100X singlet coverage per sgRNA on the Chromium V3 single-cell RNA-Seq system with Feature Barcoding (10x Genomics). Gene expression libraries were sequenced to a median depth of >50,000 reads/cell, producing a median library complexity of >3000 unique genes. Directly-captured CRISPRi sgRNAs were sequenced to >5,000 reads/cell. All Perturb-seq sequencing was performed on an Illumina NovaSeq 6000 with paired-end 100 reads.
Quantification and statistical analysis
Unified coding and lncRNA gene reference annotation
The lncRNA gene annotation corresponding to the CRiNCL library13 was merged with the ENSEMBL GRCh37.p13/hg19 annotation that was used to inform the design of the hCRISPRi libraries.22 Duplicated entries were identified using gffcompare (0.10.6). To maintain consistency across datasets, all analyses were performed using the screen feature identifiers (“feature id”) for targeted genes as the unique identifier. GTF and BED files are deposited on GitHub and a searchable database is available on the interactive web resource with direct links to the UCSC Genome Browser.
RNA-Seq processing and transcriptome analysis
RNA-Seq reads were analyzed for quality metrics using FastQC (0.11.8). Quality and adapter trimming were performed using bbduk (38.36) prior to transcript pseudoalignment and quantification using kallisto (0.45) on the unified lncRNA and coding gene annotation described above. Transcripts were aggregated to genes in R (3.6.3) using tximport (1.12.3). Expression values were processed using DESeq2 (1.24) for variance-stabilized transformation. Transcripts per million or Z-scaled variance-stabilized transformation values were used for downstream plotting and analysis. Time-course expression clustering was performed using maSigPro (1.56) at an alpha level of 0.1 using the mclust algorithm. Genes that did not significantly cluster to any of the temporal patterns were aggregated into the “unassigned” cluster.
Dual CRISPRi sgRNA libraries
Based on transcriptomic analysis, we identified expressed and dynamically regulated genes during neural induction. In order to screen equal numbers of coding and lncRNA sgRNAs, we aimed for approximately 100,000 guides for each class. Coding genes were targeted by 5 sgRNAs/locus due to well-annotated transcriptional start sites (TSS) while lncRNAs were targeted with 10 sgRNAs/locus. All sgRNAs were assembled from published CRISPRi libraries based on the human CRISPRi version 2.0 design algorithm,13,22 which uses FANTOM cap analysis of gene expression (CAGE) data to provide highly confident transcription start site coordinates. This enabled coverage of all 18,905 coding loci using the top 5 ranked sgRNAs. For lncRNA loci, we selected the combination of CRiNCL sublibraries that covered the greatest unique number of detected lncRNAs prioritized by differential expression and the temporal clustering. In total, we screened 212,938 sgRNAs targeting 29,583 loci (all 18,905 coding loci, 10,678 lncRNA loci, with 4,523 non-targeting control sgRNAs, Figure 1D).
Differentiation and proliferation screen analysis
Analysis and hit scoring of screen data was performed as previously described using ScreenProcessing.13,22 Briefly, after sgRNA quantification, all sgRNAs represented with fewer than 50 reads in any sample were excluded. Differentiation phenotypes (ρ) were calculated by taking the log2 enrichment ratio of each sgRNA in the PAX6+ versus PAX6- sorted fractions, providing a symmetric measure of the impact on neural induction (as read out by PAX6 protein) on a log-scale (Figures 1D and S1B). Proliferation phenotypes (γ) were calculated by taking the log2 ratio of the final normalized abundance versus the initial normalized abundance of each sgRNA and normalized by the number of cell divisions. For the genome-wide screen, the final abundance was calculated from the combined sum of sgRNA abundances in sorted fractions. This approach was directly validated through a separate lncRNA sublibrary screen with 37,395 sgRNAs targeting 3,560 lncRNAs (Figure S3B), where we harvested cells without sorting for the final time-point. Upon analysis, the two methods (sorted and unsorted) produced strongly correlated γ values (Pearson r = 0.99 for hits, r = 0.81 for all targets) (Figure S3B). For all analyses, a screen score that incorporated both the effect size and significance was calculated for all targets as previously described13,22,63; briefly, it is the product of the -log10 p-value and the phenotype magnitude of the top 3 sgRNAs. Hits were then identified based on this screen score, at an empirical FDR < 0.05 based on the distribution of non-targeting controls. A subset of CRiNCL sgRNAs that were within the highly active CRISPRi targeting window (1 kb around TSS) of coding genes were identified as ambiguous and excluded from all reported analyses (1468 loci); the coding loci were not excluded as they were more likely the cause of any potential phenotype. However, 142 coding loci were also excluded from analysis as they did not map to ENSEMBL hg19 annotated transcripts. Additional details on the screen scoring procedure and hit identification are previously described.13,22
Downsampling and precision-recall analysis
For estimation of hit recovery at lower levels of coverage compared to the full dataset, precision-recall analysis was performed by downsampling the raw counts data to 10%, 20%, and 50% with 1% Gaussian noise. The downsampled data then underwent the screen processing and hit identification pipeline described above. Results were compared to the full dataset for determining precision-recall and the proportion of hits recovered at each level of sampling. The median of 3 independent downsampling replicates are reported.
Gene ontology, pathway, and protein network analyses
For coding gene hits, gene ontology and KEGG pathway analyses were performed using clusterProfiler (3.14.3). Protein-protein interaction network analysis was performed using STRINGdb (11.0). For all of these analyses, we set the gene universe to contain all screened genes. For the interaction network analysis, statistical background distributions were generated through random sampling an equal number of genes from the gene universe.
Chromatin interaction analysis using MAPS
We called significant H3K4me3-mediated chromatin interactions using the MAPS pipeline64 at a resolution of 5 kb. Reads were mapped to hg19/GRCh37 using BWA-MEM (0.7.17). Unmapped reads and reads with low mapping quality were discarded. PLAC-Seq anchor bins were defined by H3K4me3 CUT&Tag using MACS2 with an q-value of 0.0001. To call significant interactions, we used a zero-truncated Poisson regression-based approach to normalize systematic biases from restriction sites, GC content, sequence repetitiveness, and ChIP enrichment. We fitted models separately for AND and XOR interactions and calculated FDRs for interactions based on the expected and observed contact frequencies between interacting 5-kb bins. We grouped interactions whose ends were located within 15 kb of each other into clusters and classified all other interactions as singletons. We defined our significant chromatin interactions as interactions with 12 or more reads, normalized contact frequency (defined as the ratio between the observed and expected contact frequency) ≥ 2, and FDR <0.01 for clusters and FDR <0.0001 for singletons. This was based on the reasoning that biologically meaningful interactions are more likely to appear in clusters, whereas singletons are more likely to represent false positives. Significant interactions were overlapped with all screened genes and annotated by hit category (e.g., lncRNA differentiation hit, coding differentiation hit, and so forth). Interactions between all combinations of categories were tallied and assessed for significance using Fisher’s exact test, with FDR-adjusted p-values for multiple testing correction.
Epigenomic dataset processing and analysis
Published neural induction epigenomics datasets were downloaded from the NIH Roadmap Epigenomics Project.29 Raw reads underwent adaptor and quality trimming using bbduk (38.36) prior to alignment using HISAT2 (2.1). For all overlap analyses, regions were counted if they overlapped by at least 1 bp within a 2 kb window centered around the transcription start site of screened genes. For quantitative analysis of ChIP-Seq signal, reads were mapped to screened genes using featureCounts 1.6 and normalized to input. For peak calling analysis of histone marks, significant peaks were identified using MACS2 (2.2.6) and replicate samples were IDR-filtered before determining overlap with screened genes following parameters of the ENCODE ChIP-Seq Pipeline for broad and narrow peaks. Genomic coordinates of enhancer regions from published datasets35,36 were downloaded as BED files and analyzed for overlap with screened genes by the same criteria. All analysis was performed on the hg19/GRCh37.p13 reference genome build, using the unified coding and lncRNA annotation described above. For visualization, bam files for replicates were merged and converted to bigwig files using deepTools 3.4.0.67 For analysis of broad H3K4me3 domains, we followed the procedure described31 by running MACS2 in with the “--broad” flag for broad peak analysis. For all genes, promoter regions (2 kb window centered around the TSS) were analyzed for overlap with H3K4me3 domains, which were categorized by percentile on their peak breadth. The top 5 percentile of peaks were assigned “broad H3K4me3” domains, as described.31
Machine learning classification
Feature data consisted of the transcriptomic and epigenomic datasets described above. Transcriptomic features included the scaled variance-stabilized and TPM values at each time-point (polyA or total RNA), log2 fold-change from day zero for each time-point, as well as variables for the maximum/median expression levels, maximum fold-change, number of exons, gene length, and isoform count. Epigenomic features consisted of the histone mark signal at the promoters (within 2 kb window surrounding the TSS) of screened genes. To prevent confounding epigenomic signal from nearby coding and lncRNA promoters, all coding-lncRNA gene pairs with promoters within the 2 kb window were excluded from classification. All predictor variables were centered around the mean and standardized. To generate machine learning models, the screen hit status was binarized and used as the response variable. For all classification models, coding genes and lncRNAs hits were compared to non-hits of the same class. For example, to analyze features of coding genes, coding hits were binarized as 1 and analyzed against coding non-hits binarized as 0. Several classes of models (elastic net logistic regression, random forest, gradient boosting machines) were generated and tested, producing similar results. Training and validation were performed using randomly-sampled partitions of 70% training data and 30% validation data. Model parameters were estimated using 5-repeats of 5-fold cross-validation. Model performance was evaluated on the validation set using the area under the receiver-operating characteristic (ROC) curve. This resampling, training, validation, and ROC assessment was repeated for 1,000 iterations, and the average AUC is reported. Each feature was additionally analyzed individually using ROC analysis to assess its association with hits. For this individual analysis, statistical significance was determined using 1,000 iterations of bootstrapping at the 99% confidence level. Variables with confidence intervals that crossed AUC 0.5 were considered non-significant.
Analysis of phenotype distribution and differentiation versus proliferation hit ratios
Differences in phenotype distributions between coding and lncRNA hits were assessed using the Kolmogorov-Smirnov (K-S) test. Skews of positive and negative phenotype distributions were assessed for significance through permutation testing. Using the baseline number of total hits for each library, we permuted the label of “positive” and “negative” hit status and calculated the ratio of positive to negative hits for 1 million trials. Skews between proliferation and differentiation hits were calculated in a similar manner, with permutation of the “proliferation” and “differentiation” hit labels performed for 1 million trials. In each case, the p-value was determined by the fraction of trials producing a more extreme ratio.
Perturb-seq computational processing
Paired-end 100 reads for gene expression and sgRNA libraries were processed using 10x cellranger software (4.0) following developer instructions for CRISPR library analysis. Data was processed on the unified lncRNA and coding gene reference described above13 as well as the newer GRCh38 genome, which led to similar results. Initial quality filtering was performed with background removal and empty droplet identification using cellbender (2.1). Barcodes identified as those belonging to cells from the cellranger and cellbender pipelines were compiled. Assignment of sgRNAs to cellular barcodes was performed using a two-component mixture model, consisting of Poisson (lower) and Gaussian (upper) distributions,18 which enabled doublet identification (barcodes with >1 sgRNA). After excluding doublets, we obtained 84,808 single cells harboring the distinct genetic perturbations. Quality scoring was performed based on unique genes detected, mitochondrial RNA percentage, and ribosomal RNA percentage. In order to assess whether apparent low-quality cellular transcriptomes were the result of any perturbations, no cells were filtered on quality metrics until after clustering and analysis (described below). After batches were integrated using multi-canonical correlation analysis in Seurat (3.9) based on the top 5000 variable genes, data was variance-stabilized and transformed using SCTransform (0.3.2). Dimensionality reduction was performed using principal components analysis followed by uniform manifold approximation projection of the top 30 principal components. High-resolution clustering of cells was performed using a shared nearest neighbor network with k = 30 and the Leiden algorithm set at a resolution of 1.2. This resulted in 30 total clusters, and clusters driven by the quality metrics described above were excluded. In total, 78,393 high-quality single-cell transcriptomes containing single sgRNA perturbations were used for all reported analysis.
Perturbation knockdown analysis
On-target knockdown of Perturb-seq targets was analyzed in a similar fashion as previously.16,18 Within each batch (10x Genomics well), cells harboring sgRNAs against each target were analyzed compared to cells harboring non-targeting control sgRNAs. To increase statistical confidence and minimize bias from gene dropout, cells were merged in a pseudobulk approach, using each batch as an individual replicate.
RNA velocity analysis
Output bam files from cellranger were processed for RNA velocity analysis of spliced an unspliced transcripts using velocyto.37 Velocity vectors were computed and visualized using scVelo,38 with 30 principal components and 30 neighbors based on the top 3000 highly variable genes for computational feasibility.
Normalized density analysis of Perturb-seq perturbations
To visualize phenotypes of Perturb-seq sgRNAs in the UMAP embedding, normalized density heatmaps of cells were constructed. For each target, cells harboring the relevant sgRNAs were identified and located in UMAP space. Gaussian kernel densities were calculated for these cells in 2 dimensions, with 10,000 total bins (100 bins in each dimension spanning the full coordinate range). To normalize for the background distribution, this density calculation was performed with the same parameters for non-targeting control sgRNAs, and this background was subtracted. The density profile was then visualized by color intensity and overlaid onto the UMAP projection.
Pairwise similarity and hierarchical clustering analysis
After calculating the normalized density for each target in the 2D UMAP embedding, density-based spatial clustering and application with noise (DBSCAN) was applied to identify areas of high density for each target. Regions of zero or near zero density were excluded using a threshold of 1% of the top The DBSCAN epsilon parameter of 1 and a minimum threshold of 25 bins were used for the top 50% of regions with highest densities. These regions were considered the enriched cell states for each target. To identify targets with similar density profiles, the overlap coefficient was determined for all pairwise comparisons and this pairwise table was converted to a distance matrix for unsupervised hierarchical clustering. To merge overlapping cell states from different targets into a universal set of cell states, all states across all targets were compared by overlap coefficient and collapsed to 29 cell states after hierarchical clustering, with the final k determined by the silhouette method for values ranging from 2 to 50.
Acknowledgments
We thank members of the Lim lab, as well as Min Cheol Kim, Rebecca Jaszczak, Bonnie Wong, Ethan Winkler, Deniz Goekbuget, Ryan Boileau, Tyler Fair, Bryan Pavlovic, Alex Pollen, and Faranak Fattahi, for helpful discussions. We thank the UCSF Parnassus Flow Cytometry Core (RRID: SCR_018206), supported in part by grant National Institutes of Health (NIH) P30 DK063720 and by the NIH S10 Instrumentation Grant S10 1S10OD021822-01. We thank Eric Chow at the UCSF Center for Advanced Technologies, funded by the Program for Breakthrough Biomedical Research Technologies, Methodologies, and Cores grant. This work was supported by NIH awards 5F31NS106868 and T32 2T32GM007618-39 to D.W. and 5R01NS091544-02, 1R01NS124881, 1R21NS101395-01, Veterans Affairs 5I01 BX000252–07, and Eli and Edythe Broad Stem Cell Innovation Pilot Award to D.A.L.
Author contributions
D.W. and D.A.L. conceptualized the study, designed experiments, and wrote the manuscript. D.W. designed, performed, and analyzed experiments and prepared figures. S.J.L., M.A.H., J.C., J.M.R., Y.S., and J.S.W. designed experiments. A.P., M.A.C., J.M.R., N.W.L.E., and G.A.C. analyzed experiments. E.H. and E.N. performed experiments. All authors reviewed and edited the manuscript.
Declaration of interests
The authors declare the following competing financial interests: The Regents of the University of California with J.S.W. and M.A.H. as inventors have filed patent applications related to CRISPRi/a screening and Perturb-seq (11,254,933). J.S.W. declares outside interest in 5AM Ventures, Amgen, Chroma Medicine, KSQ Therapeutics, Maze Therapeutics, Tenaya Therapeutics, Tessera Therapeutics, and Third Rock Ventures. M.A.H. consults for Akouos. J.M.R. consults for Maze Therapeutics.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: September 14, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2022.100177.
Supplemental information
Table of abundance (TPM) and log2 fold changes in poly(A) and total RNA fractions for all genes at multiple time-points during neural induction. Unified reference annotation (STAR Methods) available on GEO, the interactive data resource, and on the GitHub page.
Table of differentiation (ρ) and proliferation (γ) phenotypes, p values, and screen scores for all coding and lncRNA gene targets from genome-wide screens.
Table of all screened genes and their mapping to FANTOM5 atlas of human enhancers across diverse tissues and cell types and massively parallel reporter assay human neural induction enhancer dataset.
Table of all significant long-range gene-gene interactions detected by PLAC-seq.
Table of filtered singlet barcodes from Perturb-seq experiment with CRISPRi sgRNA mapping and key scRNA-seq metadata, including sgRNA protospacer sequences.
Matrix of pairwise similarity for Perturb-seq coding and lncRNA gene hits by overlap coefficient of normalized density profiles as well as tables of Perturb-seq target versus cell state (normalized density) and gene expression markers for all cell states.
Data and code availability
-
•
Sequencing data generated in this study deposited in the Gene Expression Omnibus (GEO) under accession GSE150062 and are publicly available as of the date of publication (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150062).
-
•
Custom Python scripts for analysis of genome-scale CRISPRi screens is available at https://github.com/mhorlbeck/ScreenProcessing. Custom Python scripts and Jupyter notebooks for direct capture sgRNA identity assignment are available at https://github.com/josephreplogle/guide_calling. All original code related to this work are deposited at https://github.com/symbiologist/dualgenomewide. A public, interactive R Shiny data portal that enables exploration of the collective datasets without programming experience is available at https://danlimlab.shinyapps.io/dualgenomewide. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All accession numbers and DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert S.F., Barresi M.J.F. 11th ed. Sinauer Associates; 2016. Developmental Biology. [Google Scholar]
- 4.Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M., et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perry R.B.-T., Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development. 2016;143:3882–3894. doi: 10.1242/dev.140962. [DOI] [PubMed] [Google Scholar]
- 6.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer T.R., Dinger M.E., Sunkin S.M., Mehler M.F., Mattick J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs J.A., Wolvetang E.J., Mattick J.S., Rinn J.L., Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, Plasticity, disease, and evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 9.Clark B.S., Blackshaw S. Long non coding RNA biology. Adv. Exp. Med. Biol. 2017;1008:253–282. doi: 10.1007/978-981-10-5203-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., Baker J.C., Grützner F., Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa S. Lessons from reverse-genetic studies of lncRNAs. Biochim. Biophys. Acta. 2016;1859:177–183. doi: 10.1016/j.bbagrm.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Hanna R.E., Doench J.G. Design and analysis of CRISPR–Cas experiments. Nat. Biotechnol. 2020;38:813–823. doi: 10.1038/s41587-020-0490-7. [DOI] [PubMed] [Google Scholar]
- 13.Liu S.J., Horlbeck M.A., Cho S.W., Birk H.S., Malatesta M., He D., Attenello F.J., Villalta J.E., Cho M.Y., Chen Y., et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:eaah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bester A.C., Lee J.D., Chavez A., Lee Y.-R., Nachmani D., Vora S., Victor J., Sauvageau M., Monteleone E., Rinn J.L., et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649–664.e20. doi: 10.1016/j.cell.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin N., Chang K.-Y., Li Z., Gates K., Rana Z.A., Dang J., Zhang D., Han T., Yang C.-S., Cunningham T.J., et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamson B., Norman T.M., Jost M., Cho M.Y., Nuñez J.K., Chen Y., Villalta J.E., Gilbert L.A., Horlbeck M.A., Hein M.Y., et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell. 2016;167:1867–1882.e21. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R., et al. Perturb-Seq: Dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Replogle J.M., Norman T.M., Xu A., Hussmann J.A., Chen J., Cogan J.Z., Meer E.J., Terry J.M., Riordan D.P., Srinivas N., et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol. 2020;38:954–961. doi: 10.1038/s41587-020-0470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datlinger P., Rendeiro A.F., Schmidl C., Krausgruber T., Traxler P., Klughammer J., Schuster L.C., Kuchler A., Alpar D., Bock C. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin X., Simmons S.K., Guo A., Shetty A.S., Ko M., Nguyen L., Jokhi V., Robinson E., Oyler P., Curry N., et al. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science. 2020;370:eaaz6063. doi: 10.1126/science.aaz6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tchieu J., Zimmer B., Fattahi F., Amin S., Zeltner N., Chen S., Studer L. A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell Stem Cell. 2017;21:399–410.e7. doi: 10.1016/j.stem.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horlbeck M.A., Gilbert L.A., Villalta J.E., Adamson B., Pak R.A., Chen Y., Fields A.P., Park C.Y., Corn J.E., Kampmann M., Weissman J.S. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife. 2016;5:e19760. doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parnas O., Jovanovic M., Eisenhaure T.M., Herbst R.H., Dixit A., Ye C.J., Przybylski D., Platt R.J., Tirosh I., Sanjana N.E., et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S., et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Hart T., Tong A.H.Y., Chan K., Van Leeuwen J., Seetharaman A., Aregger M., Chandrashekhar M., Hustedt N., Seth S., Noonan A., et al. Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3. 2017;7:2719–2727. doi: 10.1534/g3.117.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., Sabatini D.M. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 28.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 29.Xie W., Schultz M.D., Lister R., Hou Z., Rajagopal N., Ray P., Whitaker J.W., Tian S., Hawkins R.D., Leung D., et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C.T., Schreiber S.L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 31.Benayoun B.A., Pollina E.A., Ucar D., Mahmoudi S., Karra K., Wong E.D., Devarajan K., Daugherty A.C., Kundaje A.B., Mancini E., et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue F., Kreimer A., Ashuach T., Ahituv N., Yosef N. Identification and massively parallel characterization of regulatory elements driving neural induction. Cell Stem Cell. 2019;25:713–727.e10. doi: 10.1016/j.stem.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M.E., Lönnerberg P., Furlan A., et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J.M., Fan J., Fan H.C., Rosenfeld D., Tse D.N. An interpretable framework for clustering single-cell RNA-seq datasets. BMC Bioinformatics. 2018;19:93. doi: 10.1186/s12859-018-2092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gatchalian J., Malik S., Ho J., Lee D.-S., Kelso T.W.R., Shokhirev M.N., Dixon J.R., Hargreaves D.C. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 2018;9:5139. doi: 10.1038/s41467-018-07528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Oss S.B., Cucinotta C.E., Arndt K.M. Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem. Sci. 2017;42:788–798. doi: 10.1016/j.tibs.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J., Tam W.-L., Tong G.Q., Wu Q., Chan H.-Y., Soh B.-S., Lou Y., Yang J., Ma Y., Chai L., et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 43.Andres L.M., Blong I.W., Evans A.C., Rumachik N.G., Yamaguchi T., Pham N.D., Thompson P., Kohler J.J., Bertozzi C.R. Chemical modulation of protein O-GlcNAcylation via OGT inhibition promotes human neural cell differentiation. ACS Chem. Biol. 2017;12:2030–2039. doi: 10.1021/acschembio.7b00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jang H., Kim T.W., Yoon S., Choi S.-Y., Kang T.-W., Kim S.-Y., Kwon Y.-W., Cho E.-J., Youn H.-D. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012;11:62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Barbera J.P., Rodriguez T.A., Beddington R.S. The homeobox gene Hesx1 is required in the anterior neural ectoderm for normal forebrain formation. Dev. Biol. 2000;223:422–430. doi: 10.1006/dbio.2000.9757. [DOI] [PubMed] [Google Scholar]
- 46.Lees-Miller J.P., Heeley D.H., Smillie L.B., Kay C.M. Isolation and characterization of an abundant and novel 22-kDa protein (SM22) from chicken gizzard smooth muscle. J. Biol. Chem. 1987;262:2988–2993. [PubMed] [Google Scholar]
- 47.Tsuji-Tamura K., Morino-Koga S., Suzuki S., Ogawa M. The canonical smooth muscle cell marker TAGLN is present in endothelial cells and is involved in angiogenesis. J. Cell Sci. 2021;134 doi: 10.1242/jcs.254920. [DOI] [PubMed] [Google Scholar]
- 48.Hon C.-C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J.L., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J., et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 51.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 53.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent G., 3rd, Kenny P.J., Wahlestedt C., Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meng L., Ward A.J., Chun S., Bennett C.F., Beaudet A.L., Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barry G., Briggs J.A., Vanichkina D.P., Poth E.M., Beveridge N.J., Ratnu V.S., Nayler S.P., Nones K., Hu J., Bredy T.W., et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry. 2014;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 57.Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S., et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M., et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dimitrova N., Zamudio J.R., Jong R.M., Soukup D., Resnick R., Sarma K., Ward A.J., Raj A., Lee J.T., Sharp P.A., Jacks T. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Groff A.F., Sanchez-Gomez D.B., Soruco M.M.L., Gerhardinger C., Barutcu A.R., Li E., Elcavage L., Plana O., Sanchez L.V., Lee J.C., et al. In vivo characterization of Linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep. 2016;16:2178–2186. doi: 10.1016/j.celrep.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winkler L., Jimenez M., Zimmer J.T., Williams A., Simon M.D., Dimitrova N. Functional elements of the cis-regulatory lincRNA-p21. Cell Rep. 2022;39:110687. doi: 10.1016/j.celrep.2022.110687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mandegar M.A., Huebsch N., Frolov E.B., Shin E., Truong A., Olvera M.P., Chan A.H., Miyaoka Y., Holmes K., Spencer C.I., et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Juric I., Yu M., Abnousi A., Raviram R., Fang R., Zhao Y., Zhang Y., Qiu Y., Yang Y., Li Y., et al. MAPS: model-based analysis of long-range chromatin interactions from PLAC-seq and HiChIP experiments. PLoS Comput. Biol. 2019;15:e1006982. doi: 10.1371/journal.pcbi.1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 67.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 68.Pertea G., Pertea M. GFF Utilities: GffRead and GffCompare. F1000Res. 2020;9 doi: 10.12688/f1000research.23297.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conesa A., Nueda M.J., Ferrer A., Talón M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- 72.Kuhn M. Building predictive models in R using the caret package. J. Stat. Softw. 2008;28 [Google Scholar]
- 73.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fang R., Yu M., Li G., Chee S., Liu T., Schmitt A.D., Ren B. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 2016;26:1345–1348. doi: 10.1038/cr.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Song M., Pebworth M.-P., Yang X., Abnousi A., Fan C., Wen J., Rosen J.D., Choudhary M.N.K., Cui X., Jones I.R., et al. Cell-type-specific 3D epigenomes in the developing human cortex. Nature. 2020;587:644–649. doi: 10.1038/s41586-020-2825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table of abundance (TPM) and log2 fold changes in poly(A) and total RNA fractions for all genes at multiple time-points during neural induction. Unified reference annotation (STAR Methods) available on GEO, the interactive data resource, and on the GitHub page.
Table of differentiation (ρ) and proliferation (γ) phenotypes, p values, and screen scores for all coding and lncRNA gene targets from genome-wide screens.
Table of all screened genes and their mapping to FANTOM5 atlas of human enhancers across diverse tissues and cell types and massively parallel reporter assay human neural induction enhancer dataset.
Table of all significant long-range gene-gene interactions detected by PLAC-seq.
Table of filtered singlet barcodes from Perturb-seq experiment with CRISPRi sgRNA mapping and key scRNA-seq metadata, including sgRNA protospacer sequences.
Matrix of pairwise similarity for Perturb-seq coding and lncRNA gene hits by overlap coefficient of normalized density profiles as well as tables of Perturb-seq target versus cell state (normalized density) and gene expression markers for all cell states.
Data Availability Statement
-
•
Sequencing data generated in this study deposited in the Gene Expression Omnibus (GEO) under accession GSE150062 and are publicly available as of the date of publication (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150062).
-
•
Custom Python scripts for analysis of genome-scale CRISPRi screens is available at https://github.com/mhorlbeck/ScreenProcessing. Custom Python scripts and Jupyter notebooks for direct capture sgRNA identity assignment are available at https://github.com/josephreplogle/guide_calling. All original code related to this work are deposited at https://github.com/symbiologist/dualgenomewide. A public, interactive R Shiny data portal that enables exploration of the collective datasets without programming experience is available at https://danlimlab.shinyapps.io/dualgenomewide. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All accession numbers and DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.