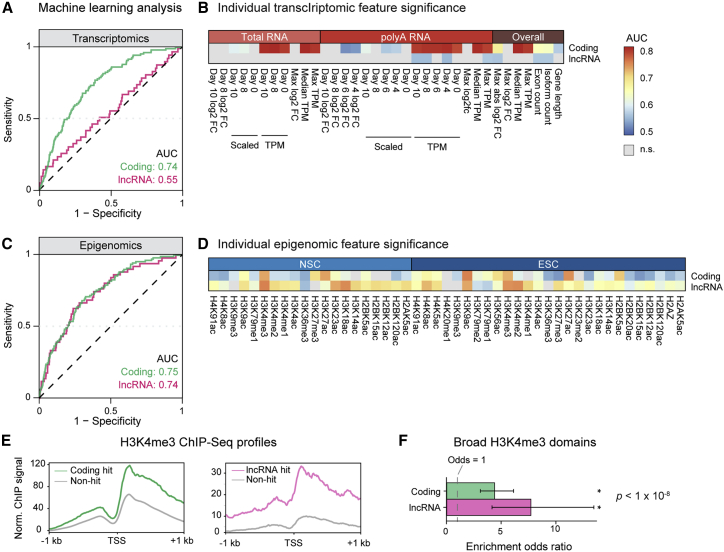
Figure 4.
Machine learning analyses reveal distinct transcriptomic and epigenomic properties of coding and lncRNA hits
(A) Representative ROC curves for transcriptomic data in classifying coding and lncRNA hits versus non-hits. Selected curves were within 1% of the mean AUC of 1,000 training/validation trials.
(B) Heatmaps showing AUC values for individual transcriptomic features for classifying coding and lncRNA hits versus non-hits. Statistical significance determined at the 99% confidence level from 1,000 bootstraps; non-significant features denoted in gray.
(C) Representative ROC curves for epigenomic data in classifying coding and lncRNA hits versus non-hits. Selected curves were within 1% of the mean AUC value of 1,000 training/validation trials.
(D) Heatmaps showing AUC values for individual epigenomic features for classifying coding and lncRNA hits versus non-hits. Statistical significance determined at the 99% confidence level from 1,000 bootstraps; non-significant features denoted in gray.
(E) ChIP-seq profiles showing average H3K4me3 signal in a 2-kb window at the promoter region of coding and lncRNA genes in ESCs. Coding hits in green, lncRNA hits in magenta, and non-hits in gray.
(F) Odds ratio for the enrichment of hits in broad H3K4me3 domains. Both coding and lncRNA gene hits were significantly enriched compared with non-hits. Dashed line denotes an odds ratio of 1 (null hypothesis), and error bars denote 95% confidence intervals by Fisher exact test. ∗p < 1 × 10−8.