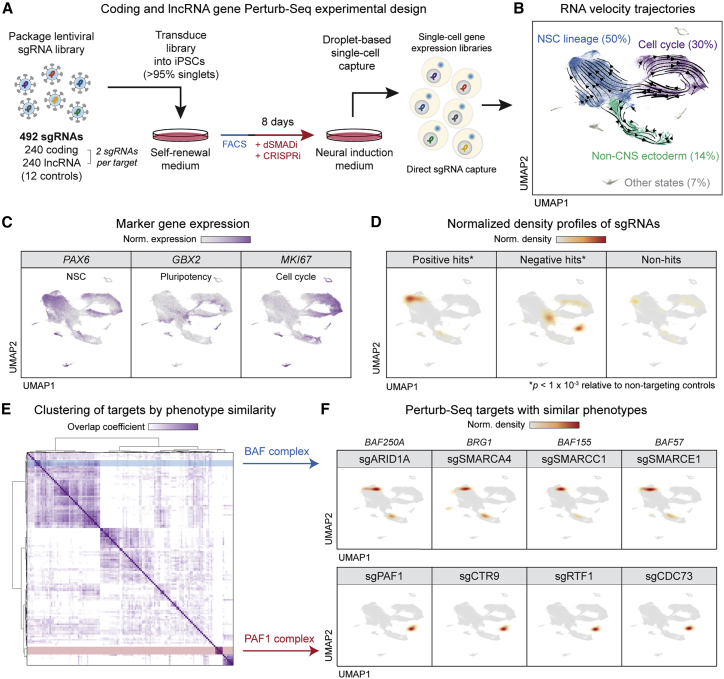
Figure 6.
Dual genome-wide screens enable Perturb-seq experiment to dissect coding and lncRNA phenotypes
(A) Overview of neural induction Perturb-seq experimental design with direct capture of sgRNAs.
(B) Major trajectories derived from RNA velocity and visualized by UMAP.
(C) Expression of markers for NSCs, pluripotent cells, and cycling cells, visualized on UMAP.
(D) Normalized density heatmaps of hit and non-hit sgRNAs on UMAP. Density profiles of target sgRNAs were calculated in UMAP space and normalized to the background density of non-targeting controls. ∗p < 1 × 10−3 based on multivariate Kolmogorov-Smirnov test versus non-targeting control distribution.
(E) Heatmap of hierarchical clustering of Perturb-seq targets by similarity of sgRNA density profiles by overlap coefficient. Two groups of targets, the BAF and PAF1 complexes, are highlighted in blue and red, respectively.
(F) Normalized density heatmaps of sgRNAs targeting BAF and PAF1 complex members, showing their colocalization in UMAP.