Abstract
Identification of circulating tumor DNA (ctDNA) following curative intent therapies is a surrogate for microscopic residual disease for patients with metastatic colorectal cancer (mCRC). Preclinically, in micrometastatic microsatellite stable (MSS) colorectal cancer, increased TGFβ signaling results in exclusion of antitumor cytotoxic T cells from the tumor microenvironment. Bintrafusp alfa (BA) is a bifunctional fusion protein composed of the extracellular domain of the TGFβRII receptor (“TGFβ trap”) and anti-PD-L1 antibody. Patients with liver-limited, MSS mCRC and with detected ctDNA after complete resection of all known tumors and standard-of-care therapy were treated with 1,200 mg of BA intravenously every 14 days for six doses. The primary endpoint was ctDNA clearance. Radiographic characteristics at recurrence were compared using independent t tests to historical data from a similar cohort of patients with liver-limited mCRC who underwent observation. Only 4 of 15 planned patients received BA before the study was stopped early for loss of equipoise. There was no grade ≥3 adverse event. None of the patients cleared ctDNA. All patients developed radiographic recurrence by the first planned restaging. Although not detectable at prior to treatment, TGFβ3 was found in circulation in all patients at cycle 2 day 1. Compared with a historical cohort, patients administered BA developed more metastases (15 vs. 2, P = 0.005) and greater tumor volumes (9 cm vs. 2 cm, P = 0.05). Treatment with BA in patients with ctDNA-detected, liver-limited mCRC did not clear ctDNA and was associated with large-volume recurrence, highlighting the potential context-specific complexity of dual TGFβ and PD-L1 inhibition.
Significance:
Use of ctDNA to identify patients with micrometastatic disease for therapeutic intervention is feasible. Treatment with BA in patients with liver-limited mCRC and with detectable ctDNA after resection generated rapid progression. Approaches targeting TGFβ signaling must consider its pathway complexity in future immunotherapy combination strategies.
Introduction
Colorectal cancer remains the second leading cause of cancer-related mortality in the United States, with approximately 50,000 deaths expected in 2021 (1). Most patients with metastatic colorectal cancer (mCRC) will develop liver metastases, which account for almost two-thirds of all colorectal cancer deaths (2). While increased use of hepatic resection (3–5) and the advent of biologic agents targeting VEGF and EGFR have improved long-term survival outcomes for the 15% to 25% of patients with mCRC who have resectable liver-limited disease, 5-year survival rates for these patients still range between 20% and 40% (6). Approximately 75% of patients with mCRC who undergo resection of liver metastases will develop disease recurrence (7). Novel approaches are therefore needed so that patients who are at high risk for recurrence can be identified earlier and offered more effective therapeutics.
Circulating tumor DNA (ctDNA) is released by tumor cells into the circulation predominantly via apoptosis (8). With a half-life on the order of hours (9), ctDNA can reveal somatic mutations harbored by tumor cells that are not present in nonmalignant cells and can serve as a “real-time” indicator of persistent cancer. The ability to detect a variant allele fraction (VAF) in the ctDNA of as low as 0.1% equips clinicians with a highly sensitive approach for identifying the presence of any microscopic foci of tumor cells (10). Indeed, in patients with colorectal cancer who undergo complete surgical resection, the detection of ctDNA is associated with inevitable disease recurrence and therefore serves as a surrogate for the existence of persistent minimal residual (micrometastatic) disease (11–15).
For the more than 95% of patients with unresectable mCRC characterized by microsatellite stability (16), immune checkpoint inhibitors targeting programmed death-1 (PD-1) and programmed death-ligand 1 (PD-L1) are ineffective, with reported overall response rates of less than 5% (17, 18). One reason for this lack of response to immunotherapy for microsatellite-stable mCRC is the absence of cytotoxic immune cells within the tumor microenvironment, which renders these tumors immunologically unreactive (19–23). In vivo, preclinical syngeneic models of colorectal cancer have demonstrated that micrometastatic tumor deposits feature activated CD4+ and CD8+ T cells that are not present in macroscopic tumors (24). In these models, this “immune exclusion” in the tumor microenvironment is mediated by upregulating signaling of the TGFβ pathway, and dual targeting of PD-L1 and TGFβ increased CD4+ and CD8+ T-cell infiltrates within the tumor microenvironment, which was not seen with inhibition of single targets alone (25).
On this basis, we hypothesized that prevention of TGFβ-induced immune exclusion along with immune checkpoint blockade may eliminate colorectal cancer micrometastases still present after resection of all evident disease. Bintrafusp alfa (BA), a bifunctional fusion protein composed of the extracellular domain of the TGFβ receptor II fused to a human IgG1 antibody blocking PD-L1, has demonstrated clinical activity and has a manageable safety profile in patients with solid tumors (26, 27). Using “ctDNA-positive” status as a surrogate for remnant micrometastatic colorectal cancer, we conducted a pilot study in patients with liver-limited mCRC who had no clinically evident disease following resection and completion of all standard-of-care therapy, to determine whether BA treatment led to clearance of ctDNA (and presumably, of micrometastatic disease).
Materials and Methods
Study Design and Participants
This trial was a prospective, single-arm pilot study of BA as monotherapy conducted under Institutional Review Board approval at The University of Texas MD Anderson Cancer Center (Houston, TX). All participants signed written informed consent prior to any study-related treatment or procedure. Patients over the age of 18 years with liver-limited metastatic adenocarcinoma of the colon or rectum who had undergone an R0 (complete) resection of their primary tumor and all known liver metastases and who had received all planned standard-of-care perioperative therapy (e.g., chemotherapy and/or radiotherapy), at the discretion of the multidisciplinary team of providers, were tested for ctDNA status (i.e., “ctDNA detected” or “not detected”). ctDNA from isolated plasma was analyzed using a 70-gene capture-based, next-generation sequencing panel of total size 150 kb (Supplementary Fig. S1) according to methodology described previously (28) and approved for use in a Clinical Laboratory Improvement Amendment (CLIA) environment. Using molecular barcoding, this panel is able to detect single-nucleotide variants in all 70 genes, copy-number variations in 19 genes, insertion-deletions in 22 genes, and fusions in six genes. ctDNA was tested at least 14 days after completion of all standard-of-care therapy. Only patients with at least one mutation detectable in the ctDNA were eligible. Only patients whose colorectal cancer had been characterized as microsatellite stable on the basis of IHC analysis showing expression of MLH1, MSH2, MSH6, and PMS2 were eligible. Patients were also required to have an Eastern Cooperative Oncology Group performance status of 0 or 1 and an estimated life expectancy exceeding 12 weeks according to the judgment of the investigator. Patients must not have had any radiographic evidence of disease at the time of study entry according to CT or MRI.
In addition, included patients must have had adequate hematologic function for study participation, defined as an absolute neutrophil count ≥1.0 × 109/L, absolute lymphocyte count ≥0.5 × 109/L, platelet count ≥100 × 109/L, and hemoglobin level ≥9.0 g/dL. In addition, patients must have had adequate renal function (defined as an estimated creatinine clearance > 30 mL/minute according to the Cockcroft-Gault formula) and adequate hepatic function [defined as a total bilirubin level ≤1.5 × the upper limit of normal (ULN), an aspartate aminotransferase level ≤2.5 × ULN, and an alanine aminotransferase level ≤2.5 × ULN].
Patients were ineligible if they had a history of extrahepatic metastases of colorectal cancer. Patients could not have had prior exposure to an immune checkpoint inhibitor or any other antineoplastic immunomodulatory agent. Patients with a history of a second primary malignancy within 3 years of study treatment were ineligible, as were patients who had undergone prior organ transplantation that necessitated ongoing immunosuppression. Patients with an active infection—including human immunodeficiency virus, hepatis B virus, hepatitis C virus, and tuberculosis—were ineligible. In addition, patients with active autoimmune disease with the potential for clinical deterioration upon treatment with BA, at the discretion of the evaluating investigator, were not allowed to participate in the study. Pregnant women were not eligible. All research conducted as a part of this clinical trial was performed in accordance with the Declaration of Helsinki, and this trial is registered at ClinicalTrials.gov (NCT03436563).
Procedures
BA (EMD Serono) was administered intravenously every 14 days at a fixed dose of 1,200 mg (Fig. 1). Toxicity was evaluated at baseline and prior to each administration of BA. Dose reductions were not permitted. Adverse events were evaluated using the NCI Common Terminology Criteria for Adverse Events (CTCAE), version 4.03 (29). Treatment with BA continued for a total of six planned doses, or until one of the following events (whichever came first): therapeutic failure requiring urgent additional antineoplastic therapy, unacceptable toxicity, onset of pregnancy, or withdrawal of informed consent.
FIGURE 1.
Study schema.
Two weeks after completion of therapy with BA, patients were evaluated for treatment response with repeat ctDNA analysis. Patients also underwent radiographic restaging studies at this time, and every 3 months thereafter, to detect any disease recurrence.
Outcomes
The primary endpoint for this study was clearance of ctDNA, defined as the disappearance of all somatic mutations identified in the blood, as well as no appearance of any new somatic mutations, following six doses of BA. Secondary endpoints were disease-free survival (DFS), calculated as the time from the date of first administration of BA until the date of documented recurrence or development of distant metastasis by RECIST (30); overall survival (OS), calculated as the time from the date of first administration of BA to the date of death by any cause; and the occurrence of grade 3 or higher adverse events according to CTCAE version 4.03.
Statistical Analysis
The planned sample size for this pilot study was 15 patients. Descriptive statistics were used to estimate the proportion of patients with ctDNA clearance, along with the associated 95% confidence interval (CI). Median DFS and OS durations (with associated 95% CIs) were estimated according to the Kaplan–Meier method (GraphPad software, version 8 was used for statistical analyses.
Circulating Biomarker Analysis
TGFβ1, 2, and 3 expression were measured in patients’ plasma, which was separated from whole blood. The other 40 soluble proteins (Supplementary Table S1) were measured in serum. Frozen plasma or serum aliquots stored in −80°C were thawed on ice immediately before performing the assay. These biomarkers were measured on the basis of multiplex electrochemiluminescence detection assays using commercially available kits from Meso Scale Discovery. The U-PLEX TGFβ Combo Human kit (K15241K-1) and V-PLEX Human Biomarker 40-Plex Kit (K15209D-1) were used. The assays were performed following the manufacturer's instructions.
Sample acquisition was performed using a QuickPlex SQ 120 instrument and analyzed using the DISCOVERY WORKBENCH 4.0 software. All samples from the same patient were run on the same plate and samples were run in technical triplicates. The results were graphed using Prism 8.0 software.
Analysis of Standard-of-care Cohort
In an unplanned, post hoc analysis, under an Institutional Review Board–approved protocol, we retrospectively reviewed databases at MD Anderson Cancer Center for patients with liver-limited mCRC in whom ctDNA was detected before surgical resection and who proceeded to observation following complete resection and standard-of-care chemotherapy. Data from these patients’ electronic medical records were obtained, including demographics, DFS, OS, vital status, and characteristics of tumor recurrence (including the number and sizes of metastases at the time of recurrence). To maintain consistency for comparison of tumor volumes across both cohorts, size dimensions of metastases were measured using the same principles for measuring target lesions according to RECIST 1.1 (31) for the purposes of estimating tumor volume. The mean number of metastases at recurrence and mean total tumor burden, defined by the sum of measurable target lesions, were compared with those for patients treated with BA using an independent t test. IBM SPSS Statistics software, version 26 was used for these analyses. Differences with a P value <0.05 were considered statistically significant.
Data Availability
All data generated in this study are available within this article and Supplementary Data.
Results
Four patients received BA on this study. As shown in Supplementary Table S2, their median age was 55.9 years (range, 42.5–68.7). Two patients had right-sided primary colon cancers, and 2 patients had sigmoid colon cancers. The median number of liver metastases at initial presentation was 4 (range, 2–4). Two patients’ tumors had KRASG12D mutations, whereas the remaining 2 patients harbored colorectal cancers expressing wild-type KRAS. All primary tumors expressed wild-type NRAS and BRAF. The median carcinoembryonic antigen (CEA) level prior to BA initiation was 4.6 ng/mL.
The median number of doses of BA received was 6 (range, 3–6). Overall, BA was tolerated well, with no grade 3 or higher treatment-related adverse events observed (Table 1). The most common adverse event was dermatitis (n = 3; all grade 1). One patient developed a keratoacanthoma of the skin, and another developed a squamous cell carcinoma of the skin. Both of these are likely related to the TGFβ trap component of the BA agent, given known linkage between disruption of TGFβ homeostasis and hyperproliferation of skin squamous epithelium (32).
TABLE 1.
Adverse events according to CTCAE, version 4.03*
| Grade 1 (No.) |
Grade 2 (No.) |
|
|---|---|---|
| Rash | 3 | 0 |
| Squamous cell carcinoma of skin | 0 | 1 |
| Actinic keratosis | 1 | 0 |
| Anorexia | 1 | 0 |
| Arthralgia | 1 | 0 |
| Condyloma | 1 | 0 |
| Congestion-nasal | 1 | 0 |
| Creatine kinase, increased | 1 | 0 |
| Diarrhea | 1 | 0 |
| Epistaxis | 1 | 0 |
| Fatigue | 1 | 0 |
| Flu-like symptoms | 1 | 0 |
| Hypothyroidism | 1 | 0 |
| Mucositis | 1 | 0 |
| Myalgia | 1 | 0 |
Abbreviations: BA, bintrafusp alfa; CEA, carcinoembryonic antigen; CI, confidence interval; CLIA, Clinical Laboratory Improvement Amendment; CRC, colorectal cancer; CTCAE, Common Terminology Criteria for Adverse Events; ctDNA, circulating tumor DNA; DFS, disease-free survival; EGFR, epidermal growth factor receptor; mCRC, metastatic colorectal cancer; OS, overall survival; PD-1, programmed death-1; PD-L1, programmed death ligand-1; TGF, transforming growth factor; ULN, upper limit of normal; VAF, variant allele fraction; VEGF, vascular endothelial growth factor.
*No grade 3 or 4 adverse events occurred.
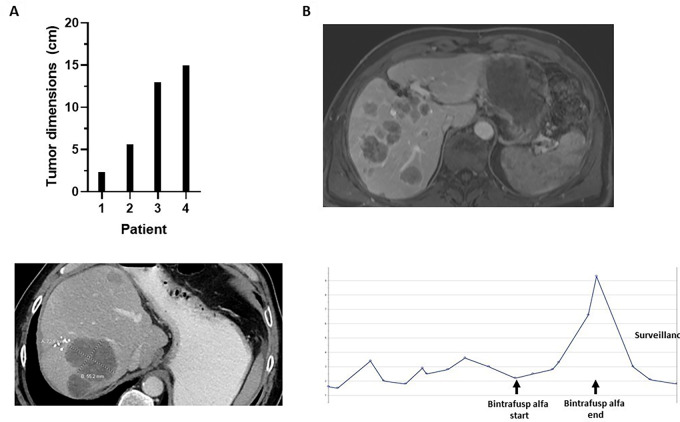
At the time of first restaging following completion of study treatment, all patients had radiographic evidence of disease recurrence (Fig. 2A). The first patient developed a single new liver metastasis measuring approximately 1 cm in its greatest diameter. The second patient developed multifocal new metastases throughout the liver, the largest measuring 7 cm in its greatest diameter. This patient also developed treatment-unrelated diverticulitis within 2 months of study discontinuation that was complicated by an extended recovery. During this time, he did not receive antineoplastic therapy for his mCRC. While off chemotherapy, his CEA level dropped from a level of 9.3 ng/mL back to within normal levels (1.5 ng/mL), and serial imaging studies showed no further growth in the size of his tumors. The third patient (Fig. 2B) developed more than 30 new liver metastases, 10 new lung metastases, and distant lymph node metastases following 3 months of treatment with BA. The fourth patient developed four new liver metastases after three doses of BA (Fig. 2C), with restaging studies conducted early at 6 weeks due to clinical suspicion of recurrence based upon a rising CEA level (Fig. 2D).
FIGURE 2.
Tumor characteristics upon recurrence after BA treatment. A, Tumor sizes. B and C, Images by CT of hepatic metastases in two different patients. D, Trend in carcinoembryonic antigen levels in one patient before, during, and after BA treatment.
The rate of ctDNA clearance following treatment with BA was 0% (95% CI, 0–60). Table 2 details the changes in ctDNA mutation profiles from baseline (before BA treatment initiation) to disease recurrence. The median number of tumor-specific mutations in the ctDNA at study enrollment was 2 (range, 1–5), with a maximum VAF of 0.5% (TP53R196* and APCA703fs) for any of the mutations detected in the pretreatment ctDNA. Three patients had plasma available for ctDNA analysis at the time of radiographic disease recurrence. In each of these patients, mutations detected before treatment were retained at recurrence, albeit often at a much higher VAF. For example, for patient 2, VAF increases were observed for APCR876* (<0.3% to 64.7%), KRASG12D (<0.3% to 60.8%), and TP53R196* (0.5% to 65.7%). Several new mutations were also observed at the time of radiographic disease recurrence. Patient 3, who only had a TP53C238Y mutation (VAF 0.3% at baseline) developed mutations in APCR1450* (VAF 23.3%), APCR216* (VAF 23.0%), KRASG12D (VAF 21.0%), SMAD4D335G (VAF 31.5%), MAPK1Q97K (VAF 1.2%), STK11D330E (VAF 0.4%), and KITR804W (VAF <0.2%) after six doses of BA.
TABLE 2.
Changes in ctDNA mutation profiles (with associated variant allele fractions) before treatment with BA and following recurrence
| Patient | Mutation | Pretreatment VAF (%) |
Postrecurrence VAF (%) |
|---|---|---|---|
| 1 | APCA703fs | 0.5 | 0.4 |
| TP53P278fs | <0.3 | 0.3 | |
| TP53C277G | 0.4 | — | |
| 2 | TP53R196* | 0.5 | 65.7 |
| APCR876* | <0.3 | 64.7 | |
| KRASG12D | <0.3 | 60.8 | |
| METN786fs | 0.3 | <0.3 | |
| MTORR206H | <0.3 | — | |
| BRCA2D1360Y | — | 0.3 | |
| 3 | TP53C238Y | 0.3 | <0.2 |
| SMAD4D335G | — | 31.5 | |
| APCR1450* | — | 23.3 | |
| APCR216* | — | 23.0 | |
| KRASG12D | — | 21.0 | |
| MAPK1Q97K | — | 1.2 | |
| STK11D330E | — | 0.4 | |
| KITR804W | — | <0.2 | |
| 4 | ERBB2R288W | <0.2 | (not tested) |
Abbreviation: VAF, variant allele fraction.
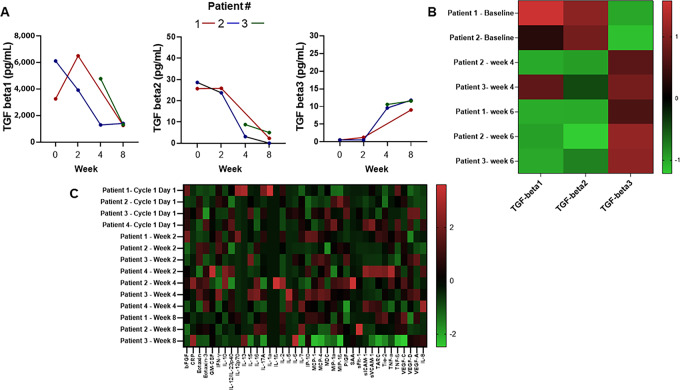
We assessed changes in cytokine and chemokines in circulation, including TGFβ isoforms. Cytokine and chemokine assessment in all patients revealed an emergence of detectable TGFβ3 in patient plasma by cycle 2, day 1 (Fig. 3A and B). However, TGFβ1 and TGFβ2 detection was reduced as expected according to the mechanism of drug action. Of the additional 40 cytokines and chemokines assessed in patient serum over time, no pattern or significant change was elsewhere observed (Fig. 3C).
FIGURE 3.
Cytokine and chemokine changes over time: TGFβ 1, 2, and 3 concentrations in plasma collected over time for individual patients (A); heatmap of normalized TGFβ 1, 2, and 3 concentrations in plasma at serial timepoints for individual participants (B); and heatmap of additional cytokines and chemokines assessed at serial timepoints in patient serum (C).
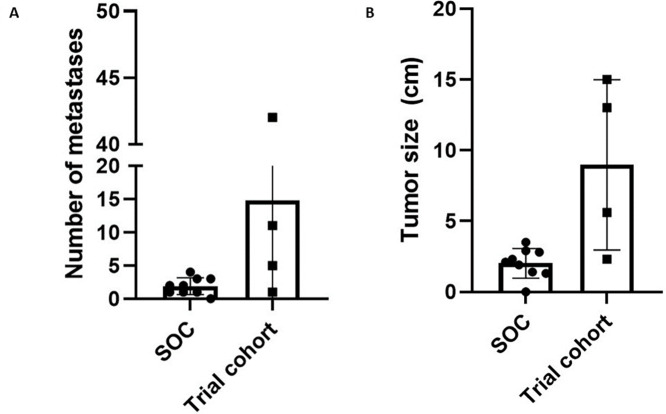
On the basis of the concerns about the rapidity and aggressiveness of the recurrences in these patients, the study team reviewed the historical recurrence patterns for patients with detectable ctDNA in patients who had previously undergone resection for liver-limited mCRC. We retrospectively identified 9 patients who had detectable ctDNA (using the same assay used for the prospective study) following completion of all standard-of-care therapy (Supplementary Table S3). There was no difference in number of liver metastases at the time of initial presentation (prior to surgery), mean size of metastases at initial presentation, age, primary tumor sidedness, or KRAS/NRAS/BRAF mutation status to suggest baseline differences in the two populations of patients (Supplementary Table S4). The mean time to the first restaging scan after completion of all planned therapies/start of observation was 3.0 months and did not differ from that of patients treated with BA (P = 0.83). While on observation, radiographic recurrence was detected in 8 of these patients. The median DFS for these patients was 4.2 months (Supplementary Fig. S2), which was somewhat longer than the median DFS of 3.0 months for patients treated with BA (HR: 4.9; 95% CI: 0.9–27.0; P = 0.07). At the time of recurrence, patients with ctDNA(+) liver-limited mCRC who proceeded to observation had a lower mean number of total metastatic lesions (2 vs. 15, P = 0.05; Fig. 4A) and smaller mean total tumor burden (2.0 cm vs. 9.0 cm, P = 0.005; Fig. 4B) than did those who received BA. Because of investigator concern that BA may be clinically detrimental, the decision was made to close the study to new patient accrual after treatment of 4 patients.
FIGURE 4.
Measurements of number of metastases (A) and tumor volume (B) for patients with liver-limited metastatic colorectal cancer who received standard-of-care (SOC) observation or BA (trial cohort).
Discussion
Here we report the clinical results from the first study, to our knowledge, to target micrometastatic colorectal cancer, informed by detection of ctDNA as a surrogate for minimal residual disease, by concomitant targeting of TGFβ signaling and PD-L1 blockade in patients with liver-limited mCRC. While BA was overall well tolerated, the rapid onset of clinical progression and the acquisition of new mutations raised concern among investigators for loss of equipoise, leading to premature discontinuation of the study.
Identification of ctDNA in the plasma following complete resection and subsequent adjuvant therapies has been demonstrated to be a biomarker of poor prognosis—a harbinger of inevitable recurrence—for patients with all stages of colorectal cancer and those with several other solid tumors (11–15, 33, 34). In one series of patients with liver-limited mCRC who underwent surgical resection of all evident disease, patients in whom ctDNA was detected in the postoperative setting had a significantly lower 2-year recurrence-free survival rate than did patients with no detectable ctDNA (0% vs. 47%, respectively; ref. 15). Consistent with this trend, our retrospective cohort of patients with liver-limited mCRC developed recurrence in most (89%) cases. Despite the strong prognostic implications of detection of ctDNA prior to eventual clinical recurrence, the clinical utility of this methodology as a predictive biomarker for response to further treatment has yet to be demonstrated, owing to a lack, thus far, of reported prospective intervention trials.
In our study, which sought to use a novel combinatory immunotherapy approach to eradicate minimal residual disease as indicated by ctDNA, the rapid onset of disease recurrence in patients with liver-limited mCRC was unexpected. Here, the 4 patients treated with BA developed higher tumor burdens, both in terms of total disease volume and the number of metastatic lesions, than was observed in a similar cohort of patients who proceeded to surveillance with no BA treatment. The occurrence of hyperprogression with immune checkpoint inhibitors has been reported in several series across solid tumors, with a prevalence of between 5% and 20% (35–37). It is possible that rapid tumor growth noted in our study was attributable to the use of anti-PD-L1 therapy and by removal of circulating TGFβ1 and TGFβ2 with the TGFβ trap of BA. However, to our surprise, all patients assessed showed an increased detection of TGFβ3 in circulation by cycle 2 day 1, a trend opposite that observed for TGFβ1 and TGFβ2. Despite significant homology in its primary structure with the other TGFβ isoforms (38), TGFβ3 is distinguished by a more “open” interaction with the TGFβ receptor II that may generate TGFβ3-specific oncogenic activation (39). While not fully characterized in colorectal cancer, inhibition of TGFβ3 in preclinical models of glioblastoma multiforme is associated with decreased expression of downstream SMAD oncogenes, decreased tumor invasiveness, and dampened TGFβ1/TGFβ2 signaling (40). The rise of circulating TGFβ3 observed in all patients following treatment with BA in our study raises concern for the possibility of a compensatory escape mechanism linked to the rapid clinical progression observed.
Interestingly, the rapid tumor growth in one patient that had been observed while on treatment with BA appeared to plateau after study drug discontinuation. The patient, whose tumor harbored a KRASG12D mutation, was monitored off systemic antineoplastic therapy for several months with minimal change, a pattern of disease biology that is not consistent with his aggressive recurrence. Murine models of KRASG12D colorectal cancer have shown that upregulation of signaling in the MAPK pathway with oncogenic KRAS mutations promotes tumor dedifferentiation for which TGFβ signaling may compensate against tumorigenesis (41). Furthermore, subsequent blocking of the type I TGFβ receptor disrupted this compensatory feedback and greatly accelerated tumor growth. This preclinical observation suggests that, in vivo, activation of MAPK signaling with impairment of prodifferentiation TGFβ activity may promote dedifferentiation and rapid development of colorectal tumors. While we were unable to compare TGFβ3 expression in the tumor tissue with that found in circulation, our observations collectively are consistent with our findings here that the treatment of micrometastatic, liver-limited mCRC with BA may have promoted tumor hyperprogression for the patients on this study.
Furthermore, TGFβ has been implicated in vivo to promote exclusion of CD4+ and CD8+ T cells within the tumor microenvironment (24). Upregulation of TGFβ signaling concurrent with growth of colorectal metastases favors creation of an immunologically inactive tumor milieu that is unresponsive to immune checkpoint blockade. Our hypothesis was that removal of TGFβ using this TGFβ trap would delay TGFβ-mediated immune suppression and render micrometastases susceptible to immune-mediated cytotoxicity driven by blockade of the PD-1–PD-L1 interaction. Unfortunately, targeting TGFβ signaling with BA in patients with micrometastatic colorectal cancer did not clear the ctDNA, nor did it promote sustained disease-free survival for these patients. In vivo, unaffected liver parenchyma harbors monocyte-derived macrophages which bind and trigger apoptosis of CD8+ T cells upon a Fas–Fas ligand interaction in our study (42). Therefore, it is possible that selection of patients in our trial specifically with liver-limited mCRC at high risk for recurrence within the liver may have been predisposed to increased hepatic clearance of T cells from the circulation that made eradication of the microscopic tumor deposits more difficult.
Our study demonstrates the feasibility of designing clinical trials seeking to eliminate micrometastatic disease, represented by ctDNA status, for patients with colorectal cancer. However, the small sample size of study participants (N = 4) limits the generalizability in drawing definitive conclusions toward concomitant targeting of the PD-1–PD-L1 axis and TGFβ signaling in the treatment of microscopic colorectal cancer. We also recognize the limitation of the small cohorts of patients represented here in making definitive comparisons of the radiographic and biochemical changes that were noted upon treatment of micrometastatic colorectal cancer with BA. The rapid increase in tumor burden upon dual targeting of PD-L1 and TGFβ and the accompanying rise in circulating TGFβ3 generate hypotheses for future study of treatment strategies of colorectal cancer which may be better informed by the observed complexity of potential compensatory signaling of multiple TGFβ isoforms (43–45).
Supplementary Material
Supplemental Table S1
Supplemental Table S2
Supplemental Table S3
Supplemental Table S4
Supplemental Figure S1
Supplemental Figure S2
Acknowledgments
The authors thank Amy Ninetto, Scientific Editor, Research Medical Library, MD Anderson Cancer Center, for editing the manuscript. V.K. Morris was supported by the NIH/NCI under award number K12 CA088084 and the Cancer Prevention & Research Institute of Texas (CPRIT) under award number RP220416. V.K. Morris, S. Kopetz, R. Luthra, and A. Dasari were supported by CPRIT under award number RP200356. J.P. Shen is a CPIRIT Scholar in Cancer Research and was supported by the NCI (L30 CA171000 and K22 CA234406), CPRIT (RR180035), the Col. Daniel Connelly Memorial Fund, and Cancer Center Support Grant (P30 CA016672). C. Haymaker was supported by the NIH/NCI under award number U24 CA224285. This study was supported by the NIH CCSG Award (CA016672 [Institutional Tissue Bank (ITB) and Research Histology Core Laboratory (RHCL)], Adaptive Patient-Oriented Longitudinal Learning and Optimization (APOLLO) Moonshot Program, Strategic Alliances and the Translational Molecular Pathology-Immunoprofiling lab (TMP-IL) at the Department Translational Molecular Pathology, the University of Texas MD Anderson Cancer Center.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
V.K. Morris reports other from EMD Serono; grants from NIH/NCI and Cancer Prevention and Research Institute of Texas (CPRIT) during the conduct of the study; other from BioNTech, Pfizer, Bristol Myers Squibb, EMD Serono, and Novartis outside the submitted work. B. Johnson reports grants from Bristol Myers Squibb, Syntrix, and Gateway for Cancer Research; personal fees from Gritstone bio, Incyte, Taiho Oncology, Insmed Oncology outside the submitted work. A. Dasari reports grants from Guardant Health, Natera, Eisai, Enterome; grants and other from HutchMed; other from Personalis and Novartis outside the submitted work. B.K. Kee reports other from Medtronic outside the submitted work. R. Huey reports personal fees from Bayer Healthcare outside the submitted work. D. Duose reports other from Chrysallis Biomedical outside the submitted work; in addition, D. Duose has a patent to ctDNA NGS panel issued. C. Haymaker reports grants from EMD Serono during the conduct of the study; other from Briacell, Mesothelioma Applied Research Foundation; personal fees from Nanobiotix and SITC; grants from Iovance, BTG, Dragonfly, Sanofi outside the submitted work. S. Kopetz reports other from MolecularMatch, Lutris, Iylon, Genentech, EMD Serono, Merck, Holy Stone, Novartis, Lilly, Boehringer Ingelheim, Boston Biomedica, AstraZeneca/MedImmune, Bayer Health, Pierre Fabre, Redx Pharma, Ipsen, Daiichi Sankyo, Natera, HalioDx, Lutris, Jacobio, Pfizer, Repare Therapeutics, Inivata, GlaxoSmithKline, Jazz Pharmaceuticals, Iylon, Xilis, Abbvie, Amal Therapeutics, Gilead Sciences, Mirati Therapeutics, Flame Biosciences, Servier, Carina Biotechnology, Bicara Therapeutics, Endeavor BioMedicines, Numab Pharma, Johnson & Johnson/Janssen, Sanofi, Biocartis, Guardant Health, Array BioPharma, Genentech/Roche, EMD Serono, MedImmune, Novartis, Amgen, Lilly, and Daiichi Sankyo during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
V.K. Morris: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing-original draft, writing-review and editing. M.J. Overman: Conceptualization, data curation, formal analysis, investigation, methodology, writing-review and editing. M. Lam: Conceptualization, resources, formal analysis, investigation, methodology, writing-original draft. C.M. Parseghian: Investigation, writing-review and editing. B. Johnson: Conceptualization, investigation, writing-review and editing. A. Dasari: Conceptualization, resources, funding acquisition, investigation, visualization, methodology, writing-review and editing. K. Raghav: Conceptualization, investigation, writing-review and editing. B.K. Kee: Investigation, writing-review and editing. R. Huey: Investigation, writing-review and editing. R.A. Wolff: Resources, funding acquisition, visualization, writing-review and editing. J.P. Shen: Investigation, writing-review and editing. J. Li: Investigation, methodology, writing-original draft, writing-review and editing. I. Zorrilla: Resources, data curation, supervision, investigation, methodology, writing-review and editing. C.-W.D. Tzeng: Investigation, writing-review and editing. H.S. Tran Cao: Investigation, writing-review and editing. Y.S. Chun: Investigation, writing-review and editing. T.E. Newhook: Investigation, writing-review and editing. N. Vauthey: Conceptualization, investigation, writing-review and editing. D. Duose: Data curation, methodology, writing-original draft, writing-review and editing. R. Luthra: Resources, investigation, methodology, writing-review and editing. C. Haymaker: Conceptualization, resources, data curation, formal analysis, supervision, investigation, methodology, writing-original draft. S. Kopetz: Conceptualization, formal analysis, supervision, investigation, visualization, methodology, writing-original draft, writing-review and editing.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2. Faivre J, Manfredi S, Bouvier AM. [Epidemiology of colorectal cancer liver metastases]. Bull Acad Natl Med 2003;187:815–22. [PubMed] [Google Scholar]
- 3. Kopetz S, Chang GJ, Overman MJ, Eng C, Sargent DJ, Larson DW, et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol 2009;27:3677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scheele J, Stang R, Altendorf-Hofmann A, Paul M. Resection of colorectal liver metastases. World J Surg 1995;19:59–71. [DOI] [PubMed] [Google Scholar]
- 5. Choti MA, Sitzmann JV, Tiburi MF, Sumetchotimetha W, Rangsin R, Schulick RD, et al. Trends in long-term survival following liver resection for hepatic colorectal metastases. Ann Surg 2002;235:759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanas GP, Taylor A, Primrose JN, Langeberg WJ, Kelsh MA, Mowat FS, et al. Survival after liver resection in metastatic colorectal cancer: review and meta-analysis of prognostic factors. Clin Epidemiol 2012;4:283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Jong MC, Pulitano C, Ribero D, Strub J, Mentha G, Schulick RD, et al. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg 2009;250:440–8. [DOI] [PubMed] [Google Scholar]
- 8. Marques JF, Junqueira-Neto S, Pinheiro J, Machado JC, Costa JL. Induction of apoptosis increases sensitivity to detect cancer mutations in plasma. Eur J Cancer 2020;127:130–8. [DOI] [PubMed] [Google Scholar]
- 9. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol 2019;5:1710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Li L, Cohen JD, Kinde I, Ptak J, Popoli M, et al. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol 2019;5:1118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reinert T, Henriksen TV, Christensen E, Sharma S, Salari R, Sethi H, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol 2019;5:1124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Overman MJ, Jaimovich A, Kennedy D, Tan M, Gavino D, Mortimer S, et al. A priori filtering of post-operative (post-op) circulating tumor DNA (ctDNA) to predict recurrence in post-metastasectomy colorectal cancer patients (CRC pts) without knowledge of tumor genotype. J Clin Oncol 36: 15s, 2018. (suppl; abstr 12044) . [Google Scholar]
- 16. Goldstein J, Tran B, Ensor J, Gibbs P, Wong HL, Wong SF, et al. Multicenter retrospective analysis of metastatic colorectal cancer (CRC) with high-level microsatellite instability (MSI-H). Ann Oncol 2014;25:1032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eng C, Kim TW, Bendell J, Argiles G, Tebbutt NC, Di Bartolomeo M, et al. Atezolizumab with or without cobimetinib versus regorafenib in previously treated metastatic colorectal cancer (IMblaze370): a multicentre, open-label, phase 3, randomised, controlled trial. Lancet Oncol 2019;20:849–61. [DOI] [PubMed] [Google Scholar]
- 19. Liu L, Mayes PA, Eastman S, Shi H, Yadavilli S, Zhang T, et al. The BRAF and MEK inhibitors dabrafenib and trametinib: effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res 2015;21:1639–51. [DOI] [PubMed] [Google Scholar]
- 20. Ebert PJR, Cheung J, Yang Y, McNamara E, Hong R, Moskalenko M, et al. MAP kinase inhibition promotes T cell and anti-tumor activity in combination with PD-L1 checkpoint blockade. Immunity 2016;44:609–21. [DOI] [PubMed] [Google Scholar]
- 21. Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016;44:698–711. [DOI] [PubMed] [Google Scholar]
- 22. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960–4. [DOI] [PubMed] [Google Scholar]
- 23. Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 2015;47:320–9. [DOI] [PubMed] [Google Scholar]
- 24. Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–43. [DOI] [PubMed] [Google Scholar]
- 25. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strauss J, Heery CR, Schlom J, Madan RA, Cao L, Kang Z, et al. Phase I trial of M7824 (MSB0011359C), a bifunctional fusion protein targeting PD-L1 and TGFbeta, in advanced solid tumors. Clin Cancer Res 2018;24:1287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahn M-J, Barlesi F, Felip E, Garon EB, Martin CM, Mok TSK, et al. Randomized open-label study of M7824 versus pembrolizumab as first-line (1L) treatment in patients with PD-L1 expressing advanced non-small cell lung cancer (NSCLC). J Clin Oncol 37: 15s, 2019. (suppl; abstr TPS9114). [Google Scholar]
- 28. Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 2015;10:e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.CTCAE version 4.03.
- 30. Seymour L, Bogaerts J, Perrone A, Ford R, Schwartz LH, Mandrekar S, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol 2017;18:e143–e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 32. Glick AB. The role of TGFbeta signaling in squamous cell cancer: lessons from mouse models. J Skin Cancer 2012;2012:249063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017;7:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oshiro C, Kagara N, Naoi Y, Shimoda M, Shimomura A, Maruyama N, et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat 2015;150:299–307. [DOI] [PubMed] [Google Scholar]
- 35. Kanjanapan Y, Day D, Wang L, Al-Sawaihey H, Abbas E, Namini A, et al. Hyperprogressive disease in early-phase immunotherapy trials: Clinical predictors and association with immune-related toxicities. Cancer 2019;125:1341–9. [DOI] [PubMed] [Google Scholar]
- 36. Matos I, Martin-Liberal J, Hierro C, Olza MOD, Viaplana C, Costa M, et al. Incidence and clinical implications of a new definition of hyperprogression (HPD) with immune checkpoint inhibitors (ICIs) in patients treated in phase 1 (Ph1) trials. J Clin Oncol 36: 15s, 2018. (suppl; abstr 3032). [Google Scholar]
- 37. Ferté C, Fernandez M, Hollebecque A, Koscielny S, Levy A, Massard C, et al. Tumor growth rate is an early indicator of antitumor drug activity in phase I clinical trials. Clin Cancer Res 2014;20:246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Crescenzo G, Hinck CS, Shu Z, Zuniga J, Yang J, Tang Y, et al. Three key residues underlie the differential affinity of the TGFbeta isoforms for the TGFbeta type II receptor. J Mol Biol 2006;355:47–62. [DOI] [PubMed] [Google Scholar]
- 39. Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP. Crystal structure of the human TbetaR2 ectodomain–TGF-beta3 complex. Nat Struct Biol 2002;9:203–8. [DOI] [PubMed] [Google Scholar]
- 40. Seystahl K, Papachristodoulou A, Burghardt I, Schneider H, Hasenbach K, Janicot M, et al. Biological role and therapeutic targeting of TGF-beta3 in glioblastoma. Mol Cancer Ther 2017;16:1177–86. [DOI] [PubMed] [Google Scholar]
- 41. Cammareri P, Vincent DF, Hodder MC, Ridgway RA, Murgia C, Nobis M, et al. TGFbeta pathway limits dedifferentiation following WNT and MAPK pathway activation to suppress intestinal tumourigenesis. Cell Death Differ 2017;24:1681–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu J, Green MD, Li S, Sun Y, Journey SN, Choi JE, et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat Med 2021;27:152–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhangu JS, Beer A, Mittlbock M, Tamandl D, Pulverer W, Schonthaler S, et al. Circulating free methylated tumor DNA markers for sensitive assessment of tumor burden and early response monitoring in patients receiving systemic chemotherapy for colorectal cancer liver metastasis. Ann Surg 2018;268:894–902. [DOI] [PubMed] [Google Scholar]
- 44. Overman MJ, Morris V, Moinova H, Manyam G, Ensor J, Lee MS, et al. Phase I/II study of azacitidine and capecitabine/oxaliplatin (CAPOX) in refractory CIMP-high metastatic colorectal cancer: evaluation of circulating methylated vimentin. Oncotarget 2016;7:67495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Osumi H, Shinozaki E, Takeda Y, Wakatsuki T, Ichimura T, Saiura A, et al. Clinical relevance of circulating tumor DNA assessed through deep sequencing in patients with metastatic colorectal cancer. Cancer Med 2019;8:408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1
Supplemental Table S2
Supplemental Table S3
Supplemental Table S4
Supplemental Figure S1
Supplemental Figure S2
Data Availability Statement
All data generated in this study are available within this article and Supplementary Data.