Figure 4.
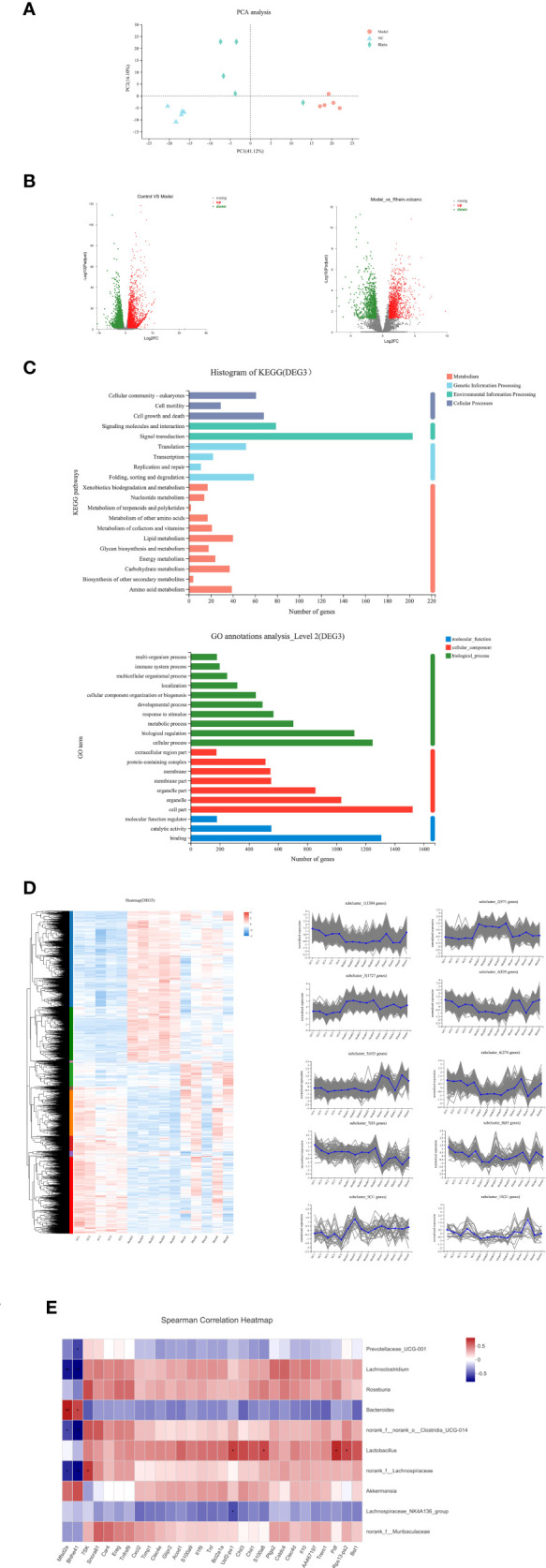
Comparisons of gene expression profiles in the mouse livers by RNA-seq among different treatment groups. (A) Principal component analysis showed that the Model group had distinct gene expression signatures compared with the NC group and rhein group PC1 (explaining 77.68% of variation). (B) Compared to the NC group, the number of differentially expressed genes (DEGs) in the Model groups was 5,220 (DEG1 2,663/2,557, upregulated and downregulated DEGs, DEGseq, respectively). Compared to the Model group, the number of differentially expressed genes (DEGs) in the rhein group was 2,503 (DEG2, 1,499/1,004, up-regulated and down-regulated DEGs, DEGseq, respectively). (C) The number of genes for Rhein vs. ALF and NC vs. ALF with the same trend was 1,907 (DEG3): KEGG annotation of DEG3 and GO annotation analysis of DEG3. (D) Heatmap of DEG3. DEG3 genes were further clustered into 10 Genes clusters according to their expression profiles. (E) Heatmap of the correlation between the altered microbial community and significantly changed gene expressions. *P < 0.05, **P < 0.01.
